Mesenchymal Stem Cell-Derived Exosomes Ameliorate Alzheimer’s Disease Pathology and Improve Cognitive Deficits
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Characterization of MSC-Exosomes
2.2. Labeling and Tracking of MSC-Exosomes
2.3. Establishment of the FAD Human Neural Cell Culture Model
2.4. Administration of MSC-Exosomes In Vitro and In Vivo
2.5. Immunoblotting
2.6. Reverse Transcription and Quantitative Real-Time Polymerase Chain Reaction (PCR)
2.7. Animals and Genotyping
2.8. Small Animal PET/CT Imaging Experimental Procedures
2.9. Novel Object Recognition (NOR) Test
2.10. Dissection of Mouse Brain
2.11. Immunofluorescence (IF) Staining
2.12. Image Acquisition and Quantification
2.13. Statistical Analysis
3. Results
3.1. Characterization of MSC-Derived Exosomes
3.2. MSC-Exosomes Decrease the Level of Aβs and Prevent the Downregulation of Neuronal Memory/Synaptic Plasticity-Related Genes
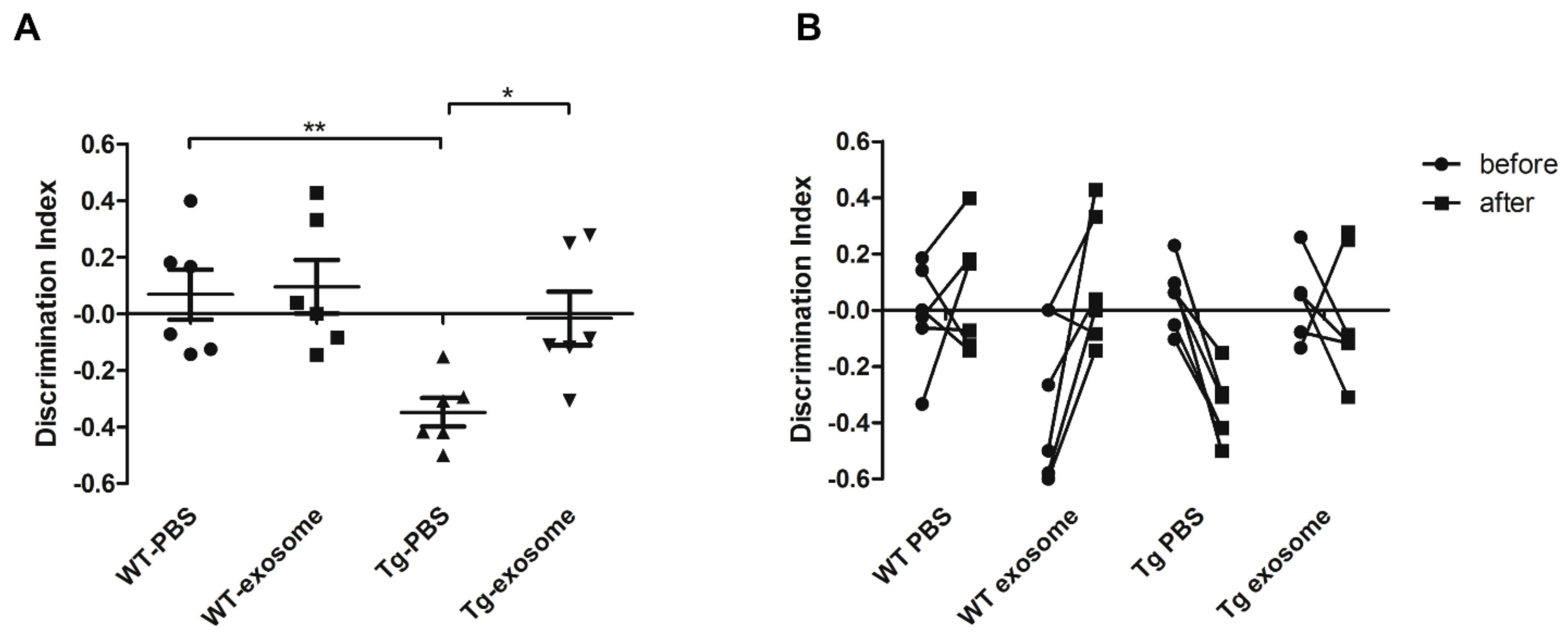
3.3. MSC-Exosomes Improve Brain Glucose Metabolism and Cognitive Function
3.4. MSC-Exosomes Reduce Aβ Plaque Burden, Inhibit Astrocyte Activation, Upregulate Neuronal Memory and Synapse-Related Genes
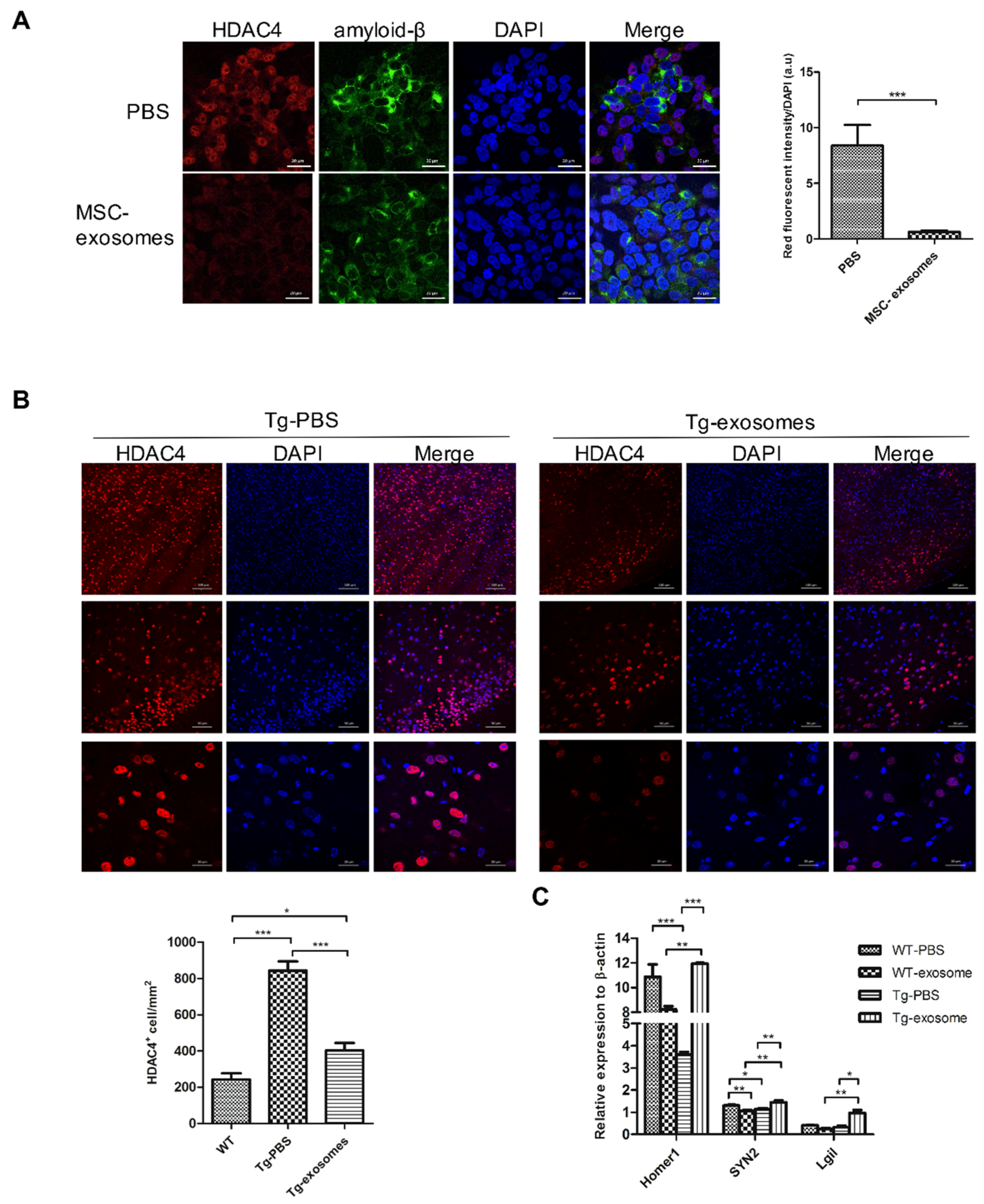
3.5. MSC-Exosomes Downregulate HDAC4 Expression and Restore Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cummings, J.; Lee, G.; Mortsdorf, T.; Ritter, A.; Zhong, K. Alzheimer’s disease drug development pipeline: 2017. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2017, 3, 367–384. [Google Scholar] [CrossRef]
- Hodson, R. Alzheimer’s disease. Nature 2018, 559, S1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, M.; Török, N.; Vécsei, L. Novel Pharmaceutical Approaches in Dementia. In NeuroPsychopharmacotherapy; Riederer, P., Laux, G., Nagatsu, T., Le, W., Riederer, C., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–18. [Google Scholar]
- Guo, T.; Zhang, D.; Zeng, Y.; Huang, T.Y.; Xu, H.; Zhao, Y. Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 40. [Google Scholar] [CrossRef]
- Herrup, K. The case for rejecting the amyloid cascade hypothesis. Nat. Neurosci. 2015, 18, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Menting, K.W.; Claassen, J.A. β-secretase inhibitor; a promising novel therapeutic drug in Alzheimer’s disease. Front. Aging Neurosci. 2014, 6, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henley, D.B.; Sundell, K.L.; Sethuraman, G.; Dowsett, S.A.; May, P.C. Safety profile of semagacestat, a gamma-secretase inhibitor: IDENTITY trial findings. Curr. Med. Res. Opin. 2014, 30, 2021–2032. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.F.; Kost, J.; Tariot, P.N.; Aisen, P.S.; Cummings, J.L.; Vellas, B.; Sur, C.; Mukai, Y.; Voss, T.; Furtek, C.; et al. Randomized Trial of Verubecestat for Mild-to-Moderate Alzheimer’s Disease. N. Engl. J. Med. 2018, 378, 1691–1703. [Google Scholar] [CrossRef]
- Van Dyck, C.H. Anti-Amyloid-beta Monoclonal Antibodies for Alzheimer’s Disease: Pitfalls and Promise. Biol. Psychiatry 2018, 83, 311–319. [Google Scholar] [CrossRef] [Green Version]
- Sevigny, J.; Chiao, P.; Bussiere, T.; Weinreb, P.H.; Williams, L.; Maier, M.; Dunstan, R.; Salloway, S.; Chen, T.; Ling, Y.; et al. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature 2016, 537, 50–56. [Google Scholar] [CrossRef]
- Huang, L.K.; Chao, S.P.; Hu, C.J. Clinical trials of new drugs for Alzheimer disease. J. Biomed. Sci. 2020, 27, 18. [Google Scholar] [CrossRef]
- Agatonovic-Kustrin, S.; Kettle, C.; Morton, D.W. A molecular approach in drug development for Alzheimer’s disease. Biomed. Pharm. 2018, 106, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.F.; Xu, T.H.; Yan, Y.; Zhou, Y.R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid beta: Structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef]
- Teixeira, F.G.; Carvalho, M.M.; Neves-Carvalho, A.; Panchalingam, K.M.; Behie, L.A.; Pinto, L.; Sousa, N.; Salgado, A.J. Secretome of mesenchymal progenitors from the umbilical cord acts as modulator of neural/glial proliferation and differentiation. Stem Cell Rev. 2015, 11, 288–297. [Google Scholar] [CrossRef]
- Zilka, N.; Zilkova, M.; Kazmerova, Z.; Sarissky, M.; Cigankova, V.; Novak, M. Mesenchymal stem cells rescue the Alzheimer’s disease cell model from cell death induced by misfolded truncated tau. Neuroscience 2011, 193, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.H.; Kim, H.N.; Park, H.J.; Shin, J.Y.; Lee, P.H. Mesenchymal Stem Cells Increase Hippocampal Neurogenesis and Neuronal Differentiation by Enhancing the Wnt Signaling Pathway in an Alzheimer’s Disease Model. Cell Transplant. 2015, 24, 1097–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivandzade, F.; Cucullo, L. Regenerative Stem Cell Therapy for Neurodegenerative Diseases: An Overview. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef]
- Phinney, D.G.; Pittenger, M.F. Concise Review: MSC-Derived Exosomes for Cell-Free Therapy. Stem Cells 2017, 35, 851–858. [Google Scholar] [CrossRef] [Green Version]
- Mendt, M.; Rezvani, K.; Shpall, E. Mesenchymal stem cell-derived exosomes for clinical use. Bone Marrow Transplant. 2019, 54, 789–792. [Google Scholar] [CrossRef]
- Pant, S.; Hilton, H.; Burczynski, M.E. The multifaceted exosome: Biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem. Pharmacol. 2012, 83, 1484–1494. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, J.; Stewart, T.; Sheng, L.; Li, N.; Bullock, K.; Song, N.; Shi, M.; Banks, W.A.; Zhang, J. Transmission of alpha-synuclein-containing erythrocyte-derived extracellular vesicles across the blood-brain barrier via adsorptive mediated transcytosis: Another mechanism for initiation and progression of Parkinson’s disease? Acta Neuropathol. Commun. 2017, 5, 71. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.C.; Liu, L.; Ma, F.; Wong, C.W.; Guo, X.E.; Chacko, J.V.; Farhoodi, H.P.; Zhang, S.X.; Zimak, J.; Segaliny, A.; et al. Elucidation of Exosome Migration across the Blood-Brain Barrier Model In Vitro. Cell. Mol. Bioeng. 2016, 9, 509–529. [Google Scholar] [CrossRef]
- Cui, G.H.; Wu, J.; Mou, F.F.; Xie, W.H.; Wang, F.B.; Wang, Q.L.; Fang, J.; Xu, Y.W.; Dong, Y.R.; Liu, J.R.; et al. Exosomes derived from hypoxia-preconditioned mesenchymal stromal cells ameliorate cognitive decline by rescuing synaptic dysfunction and regulating inflammatory responses in APP/PS1 mice. FASEB J. 2018, 32, 654–668. [Google Scholar] [CrossRef] [Green Version]
- Reza-Zaldivar, E.E.; Hernandez-Sapiens, M.A.; Gutierrez-Mercado, Y.K.; Sandoval-Avila, S.; Gomez-Pinedo, U.; Marquez-Aguirre, A.L.; Vazquez-Mendez, E.; Padilla-Camberos, E.; Canales-Aguirre, A.A. Mesenchymal stem cell-derived exosomes promote neurogenesis and cognitive function recovery in a mouse model of Alzheimer’s disease. Neural Regen. Res. 2019, 14, 1626–1634. [Google Scholar] [CrossRef]
- Reza-Zaldivar, E.E.; Hernandez-Sapiens, M.A.; Minjarez, B.; Gutierrez-Mercado, Y.K.; Marquez-Aguirre, A.L.; Canales-Aguirre, A.A. Potential Effects of MSC-Derived Exosomes in Neuroplasticity in Alzheimer’s Disease. Front. Cell. Neurosci. 2018, 12, 317. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Kim, Y.H.; Hebisch, M.; Sliwinski, C.; Lee, S.; D’Avanzo, C.; Chen, H.; Hooli, B.; Asselin, C.; Muffat, J.; et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 2014, 515, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.; Politis, M.; Rabiner, E.A.; Middleton, L.T. Novel PET Biomarkers to Disentangle Molecular Pathways across Age-Related Neurodegenerative Diseases. Cells 2020, 9, 2581. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.P.; Mardini, O.; Ericsson, M.; Prabhakar, S.; Maguire, C.; Chen, J.W.; Tannous, B.A.; Breakefield, X.O. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano 2014, 8, 483–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.H.; Choi, S.H.; D’Avanzo, C.; Hebisch, M.; Sliwinski, C.; Bylykbashi, E.; Washicosky, K.J.; Klee, J.B.; Brustle, O.; Tanzi, R.E.; et al. A 3D human neural cell culture system for modeling Alzheimer’s disease. Nat. Protoc. 2015, 10, 985–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsuda, T.; Tsuchiya, R.; Kosaka, N.; Yoshioka, Y.; Takagaki, K.; Oki, K.; Takeshita, F.; Sakai, Y.; Kuroda, M.; Ochiya, T. Human adipose tissue-derived mesenchymal stem cells secrete functional neprilysin-bound exosomes. Sci. Rep. 2013, 3, 1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahim, A.G.; Cheng, K.; Marban, E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Rep. 2014, 2, 606–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chopp, M.; Liu, X.S.; Katakowski, M.; Wang, X.; Tian, X.; Wu, D.; Zhang, Z.G. Exosomes Derived from Mesenchymal Stromal Cells Promote Axonal Growth of Cortical Neurons. Mol. Neurobiol. 2017, 54, 2659–2673. [Google Scholar] [CrossRef]
- Hu, Y.; Rao, S.S.; Wang, Z.X.; Cao, J.; Tan, Y.J.; Luo, J.; Li, H.M.; Zhang, W.S.; Chen, C.Y.; Xie, H. Exosomes from human umbilical cord blood accelerate cutaneous wound healing through miR-21-3p-mediated promotion of angiogenesis and fibroblast function. Theranostics 2018, 8, 169–184. [Google Scholar] [CrossRef]
- He, L.; Zhu, W.; Chen, Q.; Yuan, Y.; Wang, Y.; Wang, J.; Wu, X. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics 2019, 9, 8206–8220. [Google Scholar] [CrossRef]
- Choi, H.; Lee, D.S. Illuminating the physiology of extracellular vesicles. Stem Cell Res. Ther. 2016, 7, 55. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginsberg, S.D.; Alldred, M.J.; Che, S. Gene expression levels assessed by CA1 pyramidal neuron and regional hippocampal dissections in Alzheimer’s disease. Neurobiol. Dis. 2012, 45, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Karl, T.; Bhatia, S.; Cheng, D.; Kim, W.S.; Garner, B. Cognitive phenotyping of amyloid precursor protein transgenic J20 mice. Behav. Brain Res. 2012, 228, 392–397. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Chen, J.; Li, J.; Kofler, J.; Herrup, K. Neurons in Vulnerable Regions of the Alzheimer’s Disease Brain Display Reduced ATM Signaling. Eneuro 2016, 3. [Google Scholar] [CrossRef] [Green Version]
- Anderson, K.W.; Chen, J.; Wang, M.; Mast, N.; Pikuleva, I.A.; Turko, I.V. Quantification of histone deacetylase isoforms in human frontal cortex, human retina, and mouse brain. PLoS ONE 2015, 10, e0126592. [Google Scholar] [CrossRef] [Green Version]
- Tarasoff-Conway, J.M.; Carare, R.O.; Osorio, R.S.; Glodzik, L.; Butler, T.; Fieremans, E.; Axel, L.; Rusinek, H.; Nicholson, C.; Zlokovic, B.V.; et al. Clearance systems in the brain-implications for Alzheimer disease. Nat. Rev. Neurol. 2015, 11, 457–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Cui, G.H.; Guo, H.D.; Li, H.; Zhai, Y.; Gong, Z.B.; Wu, J.; Liu, J.S.; Dong, Y.R.; Hou, S.X.; Liu, J.R. RVG-modified exosomes derived from mesenchymal stem cells rescue memory deficits by regulating inflammatory responses in a mouse model of Alzheimer’s disease. Immun. Ageing 2019, 16, 10. [Google Scholar] [CrossRef] [Green Version]
- Pardridge, W.M. Drug transport across the blood-brain barrier. J. Cereb. Blood Flow Metab. 2012, 32, 1959–1972. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.F.; Herholz, K.; Rosa-Neto, P.; Pellerin, L.; Nordberg, A.; Zimmer, E.R. Astrocyte Biomarkers in Alzheimer’s Disease. Trends Mol. Med. 2019, 25, 77–95. [Google Scholar] [CrossRef]
- Stoessl, A.J. Glucose utilization: Still in the synapse. Nat. Neurosci. 2017, 20, 382–384. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, E.R.; Parent, M.J.; Souza, D.G.; Leuzy, A.; Lecrux, C.; Kim, H.I.; Gauthier, S.; Pellerin, L.; Hamel, E.; Rosa-Neto, P. [(18)F]FDG PET signal is driven by astroglial glutamate transport. Nat. Neurosci. 2017, 20, 393–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heneka, M.T.; Sastre, M.; Dumitrescu-Ozimek, L.; Dewachter, I.; Walter, J.; Klockgether, T.; Van Leuven, F. Focal glial activation coincides with increased BACE1 activation and precedes amyloid plaque deposition in APP[V717I] transgenic mice. J. Neuroinflamm. 2005, 2, 22. [Google Scholar] [CrossRef] [Green Version]
- Kraft, A.W.; Hu, X.; Yoon, H.; Yan, P.; Xiao, Q.; Wang, Y.; Gil, S.C.; Brown, J.; Wilhelmsson, U.; Restivo, J.L.; et al. Attenuating astrocyte activation accelerates plaque pathogenesis in APP/PS1 mice. FASEB J. 2013, 27, 187–198. [Google Scholar] [CrossRef] [Green Version]
- McKiernan, E.F.; O’Brien, J.T. 7T MRI for neurodegenerative dementias in vivo: A systematic review of the literature. J. Neurol. Neurosurg. Psychiatry 2017, 88, 564–574. [Google Scholar] [CrossRef] [Green Version]
- Burda, J.E.; Sofroniew, M.V. Reactive gliosis and the multicellular response to CNS damage and disease. Neuron 2014, 81, 229–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, S.F.; Scholl, M.; Almkvist, O.; Wall, A.; Engler, H.; Langstrom, B.; Nordberg, A. Evidence for astrocytosis in prodromal Alzheimer disease provided by 11C-deuterium-L-deprenyl: A multitracer PET paradigm combining 11C-Pittsburgh compound B and 18F-FDG. J. Nuclear Med. 2012, 53, 37–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orr, A.G.; Hsiao, E.C.; Wang, M.M.; Ho, K.; Kim, D.H.; Wang, X.; Guo, W.; Kang, J.; Yu, G.Q.; Adame, A.; et al. Astrocytic adenosine receptor A2A and Gs-coupled signaling regulate memory. Nat. Neurosci. 2015, 18, 423–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, M.; Yin, Z.; Chen, F.; Lei, P. Mesenchymal stem cell-derived exosome: A promising alternative in the therapy of Alzheimer’s disease. Alzheimer’s Res. Ther. 2020, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Sala Frigerio, C.; Wolfs, L.; Fattorelli, N.; Thrupp, N.; Voytyuk, I.; Schmidt, I.; Mancuso, R.; Chen, W.T.; Woodbury, M.E.; Srivastava, G.; et al. The Major Risk Factors for Alzheimer’s Disease: Age, Sex, and Genes Modulate the Microglia Response to Aβ Plaques. Cell Rep. 2019, 27, 1293–1306.e6. [Google Scholar] [CrossRef] [Green Version]
- Castanho, I.; Murray, T.K.; Hannon, E.; Jeffries, A.; Walker, E.; Laing, E.; Baulf, H.; Harvey, J.; Bradshaw, L.; Randall, A.; et al. Transcriptional Signatures of Tau and Amyloid Neuropathology. Cell Rep. 2020, 30, 2040–2054.e5. [Google Scholar] [CrossRef] [Green Version]
- Sierksma, A.; Lu, A.; Mancuso, R.; Fattorelli, N.; Thrupp, N.; Salta, E.; Zoco, J.; Blum, D.; Buee, L.; De Strooper, B.; et al. Novel Alzheimer risk genes determine the microglia response to amyloid-beta but not to TAU pathology. EMBO Mol. Med. 2020, 12, e10606. [Google Scholar] [CrossRef]
- Yang, H.S.; Onos, K.D.; Choi, K.; Keezer, K.J.; Skelly, D.A.; Carter, G.W.; Howell, G.R. Natural genetic variation determines microglia heterogeneity in wild-derived mouse models of Alzheimer’s disease. Cell Rep. 2021, 34, 108739. [Google Scholar] [CrossRef]
- Kota, J.; Chivukula, R.R.; O’Donnell, K.A.; Wentzel, E.A.; Montgomery, C.L.; Hwang, H.W.; Chang, T.C.; Vivekanandan, P.; Torbenson, M.; Clark, K.R.; et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009, 137, 1005–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, C.K.; Jena, B.C.; Banerjee, I.; Das, S.; Parekh, A.; Bhutia, S.K.; Mandal, M. Exosome as a Novel Shuttle for Delivery of Therapeutics across Biological Barriers. Mol. Pharm. 2019, 16, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Penney, J.; Tsai, L.H. Histone deacetylases in memory and cognition. Sci. Signal. 2014, 7, re12. [Google Scholar] [CrossRef] [PubMed]
- Graff, J.; Rei, D.; Guan, J.S.; Wang, W.Y.; Seo, J.; Hennig, K.M.; Nieland, T.J.; Fass, D.M.; Kao, P.F.; Kahn, M.; et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 2012, 483, 222–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haberland, M.; Montgomery, R.L.; Olson, E.N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009, 10, 32–42. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-A.; Lu, C.-H.; Ke, C.-C.; Chiu, S.-J.; Jeng, F.-S.; Chang, C.-W.; Yang, B.-H.; Liu, R.-S. Mesenchymal Stem Cell-Derived Exosomes Ameliorate Alzheimer’s Disease Pathology and Improve Cognitive Deficits. Biomedicines 2021, 9, 594. https://doi.org/10.3390/biomedicines9060594
Chen Y-A, Lu C-H, Ke C-C, Chiu S-J, Jeng F-S, Chang C-W, Yang B-H, Liu R-S. Mesenchymal Stem Cell-Derived Exosomes Ameliorate Alzheimer’s Disease Pathology and Improve Cognitive Deficits. Biomedicines. 2021; 9(6):594. https://doi.org/10.3390/biomedicines9060594
Chicago/Turabian StyleChen, Yi-An, Cheng-Hsiu Lu, Chien-Chih Ke, Sain-Jhih Chiu, Fong-Shya Jeng, Chi-Wei Chang, Bang-Hung Yang, and Ren-Shyan Liu. 2021. "Mesenchymal Stem Cell-Derived Exosomes Ameliorate Alzheimer’s Disease Pathology and Improve Cognitive Deficits" Biomedicines 9, no. 6: 594. https://doi.org/10.3390/biomedicines9060594
APA StyleChen, Y.-A., Lu, C.-H., Ke, C.-C., Chiu, S.-J., Jeng, F.-S., Chang, C.-W., Yang, B.-H., & Liu, R.-S. (2021). Mesenchymal Stem Cell-Derived Exosomes Ameliorate Alzheimer’s Disease Pathology and Improve Cognitive Deficits. Biomedicines, 9(6), 594. https://doi.org/10.3390/biomedicines9060594






