Vocal Fold Fibroblasts in Reinke’s Edema Show Alterations Involved in Extracellular Matrix Production, Cytokine Response and Cell Cycle Control
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Tissue Samples
2.2. Cell Culture
2.3. Proliferation Assay
2.4. Sample Collection
2.4.1. Supernatant for Luminex® Assays, ELISAs and Silver Stain
2.4.2. RNA Isolation
2.4.3. Cell lysates for Protein Analysis
2.5. Sample Analysis
2.5.1. RNA-seq and Enrichment Analysis
2.5.2. Reverse Transcription (RT) and Quantitative Polymerase Chain Reaction (qPCR)
2.5.3. SDS-PAGE and Western Blot Analysis
2.5.4. Pepsin Digestion, SDS-PAGE and Silver Stain
2.5.5. ELISA
2.5.6. Magnetic Luminex® Assay
2.6. Statistical Analysis
3. Results
3.1. RNA-Seq Reveals a Total of 74 Differentially Expressed Genes in RE VFF Cells
3.2. Gene Expression of AQP1, and VWF, But Not Other Endothelial Cell Markers, Is Significantly Upregulated in RE VFF
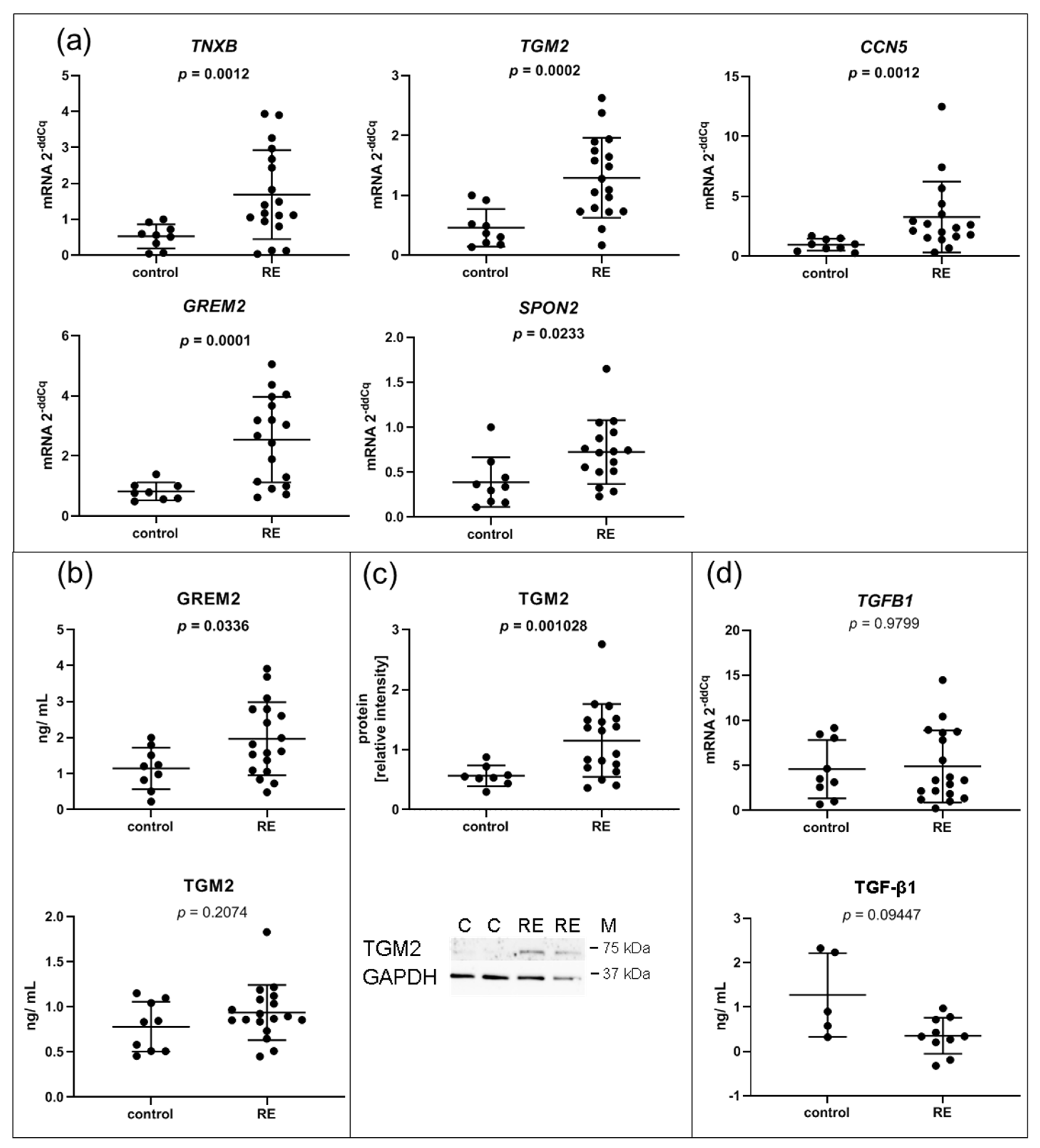
3.3. Expression of Genes Associated with the ECM Composition, Some of Which Involved in the Interaction with TGF-β Family Members, Are Significantly Upregulated in RE VFF
3.4. MMP1 Is Decreased in RE VFF Leading to Accumulation of COL1α2
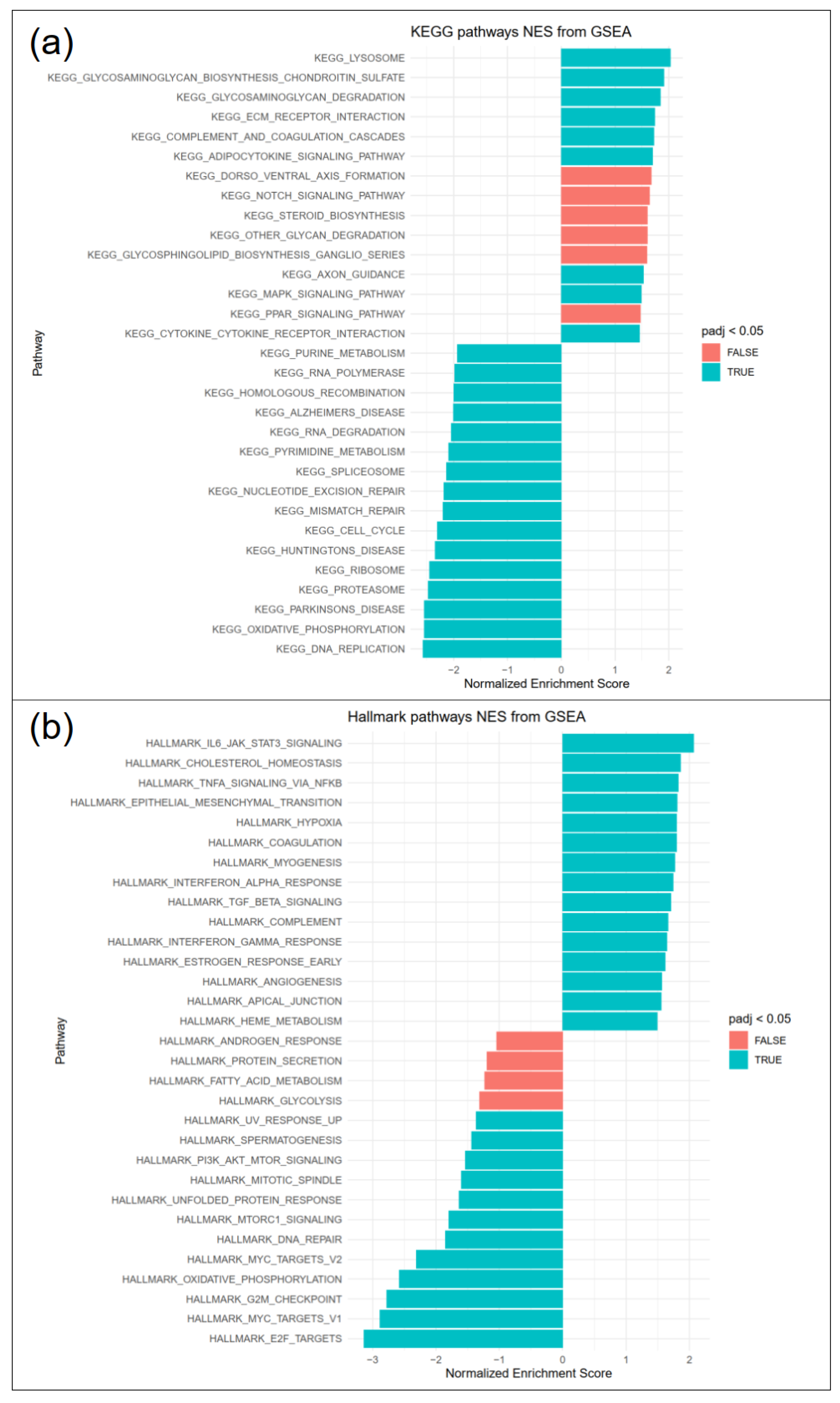
3.5. Pathways Involved in ECM Interactions and Turnover, Inflammation and Fibrosis Are Upregulated in RE, While Cell Proliferation Is Downregulated
3.6. RE VFFs Have Increased Levels of Inflammatory Cytokine Receptors IL1R1 and IL6R
3.7. Cell Cycle, DNA Replication and E2F Targets Are Downregulated in RE VFF
4. Discussion
4.1. Cultured RE VFF Reveal Alterations Associated with ECM Remodeling, Fibrosis and Angiogenesis
4.2. RE VFF Display Changes Leading to A Differential Inflammatory Response
4.3. RE VFF Show Cell Cycle Disruption
4.4. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krischke, S.; Weigelt, S.; Hoppe, U.; Köllner, V.; Klotz, M.; Eysholdt, U.; Rosanowski, F. Quality of Life in Dysphonic Patients. J. Voice 2005, 19, 132–137. [Google Scholar] [CrossRef]
- Rosow, D.E.; Szczupak, M.; Saint-Victor, S.; Gerhard, J.D.; Dupont, C.; Lo, K. The economic impact of vocal attrition in public school teachers in Miami-Dade County. Laryngoscope 2015, 126, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Tavaluc, R.; Tan-Geller, M. Reinke’s Edema. Otolaryngol. Clin. N. Am. 2019, 52, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Dikkers, F.G.; Nikkels, P.G.J. Benign Lesions of the Vocal Folds: Histopathology and Phonotrauma. Ann. Otol. Rhinol. Laryngol. 1995, 104, 698–703. [Google Scholar] [CrossRef]
- Lim, J.-Y.; Choi, J.-N.; Kim, K.-M.; Choi, H.-S. Voice analysis of patients with diverse types of Reinke’s edema and clinical use of electroglottographic measurements. Acta Oto Laryngol. 2006, 126, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Marcotullio, D.; Magliulo, G.; Pezone, T. Reinke’s edema and risk factors: Clinical and histopathologic aspects. Am. J. Otolaryngol. 2002, 23, 81–84. [Google Scholar] [CrossRef]
- Branski, R.C.; Saltman, B.; Sulica, L.; Szeto, H.; Duflo, S.; Felsen, D.; Kraus, D.H. Cigarette smoke and reactive oxygen species metabolism: Implications for the pathophysiology of Reinke’s edema. Laryngoscope 2009, 119, 2014–2018. [Google Scholar] [CrossRef]
- Sato, K.; Hirano, M.; Nakashima, T. Electron microscopic and immunohistochemical investigation of Reinke’s edema. Ann. Otol. Rhinol. Laryngol. 1999, 108, 1068–1072. [Google Scholar] [CrossRef]
- Martins, R.H.G.; Tavares, E.L.M.; Pessin, A.B.B. Are Vocal Alterations Caused by Smoking in Reinke’s Edema in Women Entirely Reversible After Microsurgery and Smoking Cessation? J. Voice 2017, 31, 380.e11–380.e14. [Google Scholar] [CrossRef] [Green Version]
- Jovanovic, M.B.; Mulutinovic, Z.; Perovic, J.; Grubor, A.; Milenković, S.; Malobabić, S. Contact Telescopy Reveals Blood Vessel Alterations of Vocal Fold Mucosa in Reinke’s Edema. J. Voice 2007, 21, 355–360. [Google Scholar] [CrossRef]
- Foote, A.G.; Wang, Z.; Kendziorski, C.; Thibeault, S.L. Tissue specific human fibroblast differential expression based on RNAsequencing analysis. BMC Genom. 2019, 20, 308. [Google Scholar] [CrossRef] [PubMed]
- Gugatschka, M.; Darnhofer, B.; Grossmann, T.; Schittmayer, M.; Hortobagyi, D.; Kirsch, A.; Karpf, E.; Brcic, L.; Birner-Gruenberger, R.; Karbiener, M. Proteomic Analysis of Vocal Fold Fibroblasts Exposed to Cigarette Smoke Extract: Exploring the Pathophysiology of Reinke’s Edema *. Mol. Cell. Proteom. 2019, 18, 1511–1525. [Google Scholar] [CrossRef]
- van Linthout, S.; Miteva, K.; Tschöpe, C. Crosstalk between fibroblasts and inflammatory cells. Cardiovasc. Res. 2014, 102, 258–269. [Google Scholar] [CrossRef] [Green Version]
- Duflo, S.M.; Thibeault, S.L.; Li, W.; Smith, M.E.; Schade, G.; Hess, M.M. Differential gene expression profiling of vocal fold polyps and Reinke’s edema by complementary DNA microarray. Ann. Otol. Rhinol. Laryngol. 2006, 115, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Graupp, M.; Rinner, B.; Frisch, M.T.; Weiss, G.; Fuchs, J.; Sundl, M.; El-Heliebi, A.; Moser, G.; Kamolz, L.P.; Karbiener, M.; et al. Towards an in vitro fibrogenesis model of human vocal fold scarring. Eur. Arch. Oto Rhino Laryngol. 2018, 275, 1211–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Chapman, B.A.; Kirchner, R.D.; Pantano, L.; Naumenko, S.; de Smet, M.; Beltrame, L.; Khotiainsteva, T.; Sytchev, I.; Guimera, R.V.; Saveliev, V.; et al. Bcbio/Bcbio-Nextgen: V1.1.5. Available online: https://zenodo.org/record/4556385/export/json#.YNV_26hLg2w (accessed on 23 June 2021).
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of Samtools and Bcftools. Gigascience 2021, 10. [Google Scholar] [CrossRef]
- Didion, J.P.; Martin, M.; Collins, F.S. Atropos: Specific, Sensitive, and Speedy Trimming of Sequencing Reads. PeerJ. 2017, 5, e3720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. Version 4.0.3. R Foundation for Statistical Computing. 2020. Available online: http://www.R-project.org/ (accessed on 23 June 2021).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Steinbaugh, M.J.; Pantano, L.; Kirchner, R.D.; Barrera, V.; Chapman, B.A.; Piper, M.E.; Mistry, M.; Khetani, R.S.; Rutherford, K.D.; Hofmann, O.; et al. bcbioRNASeq: R package for bcbio RNA-seq analysis. F1000Research 2018, 6, 1976. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast Gene Set Enrichment Analysis. bioRxiv 2021. Available online: https://www.biorxiv.org/content/10.1101/060012v3 (accessed on 23 June 2021). [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, T.; Steffan, B.; Kirsch, A.; Grill, M.; Msc, C.G.; Gugatschka, M. Exploring the Pathophysiology of Reinke’s Edema: The Cellular Impact of Cigarette Smoke and Vibration. Laryngoscope 2021, 131, E547–E554. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.D.; Titze, I.R.; Chan, R.; Bs, T.H.H. Vocal fold proteoglycans and their influence on biomechanics. Laryngoscope 1999, 109, 845–854. [Google Scholar] [CrossRef]
- Verkman, A.S.; Anderson, M.O.; Papadopoulos, M. Aquaporins: Important but elusive drug targets. Nat. Rev. Drug Discov. 2014, 13, 259–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altuntas, A.; Yilmaz, M.D.; Aktepe, F.; Kahveci, O.K.; Derekoy, S.; Dilek, H.; Serteser, M. Expression and distribution of aquaporin-1 in nasal polyps: Does it have any significance in edema formation? Am. J. Rhinol. 2006, 20, 128–131. [Google Scholar] [CrossRef]
- Medici, D.; Kalluri, R. Endothelial–mesenchymal transition and its contribution to the emergence of stem cell phenotype. Semin. Cancer Biol. 2012, 22, 379–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nurminskaya, M.V.; Belkin, A.M. Cellular Functions of Tissue Transglutaminase. Int. Rev. Cell Mol. Biol. 2012, 294, 1–97. [Google Scholar] [CrossRef] [Green Version]
- Kojima, S.; Nara, K.; Rifkin, D.B. Requirement for transglutaminase in the activation of latent transforming growth factor-beta in bovine endothelial cells. J. Cell Biol. 1993, 121, 439–448. [Google Scholar] [CrossRef]
- Wang, Z.; Perez, M.; Lee, E.-S.; Kojima, S.; Griffin, M. The functional relationship between transglutaminase 2 and transforming growth factor β1 in the regulation of angiogenesis and endothelial–mesenchymal transition. Cell Death Dis. 2017, 8, e3032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirza, A.; Liu, S.L.; Frizell, E.; Zhu, J.; Maddukuri, S.; Martinez, J.; Davies, P.; Schwarting, R.; Norton, P.; Zern, M.A. A role for tissue transglutaminase in hepatic injury and fibrogenesis, and its regulation by NF-kappaB. Am. J. Physiol. Liver Physiol. 1997, 272, G281–G288. [Google Scholar] [CrossRef]
- Suto, N.; Ikura, K.; Sasaki, R. Expression induced by interleukin-6 of tissue-type transglutaminase in human hepatoblastoma HepG2 cells. J. Biol. Chem. 1993, 268, 7469–7473. [Google Scholar] [CrossRef]
- Ritter, S.J.; Davies, P.J.A. Identification of a Transforming Growth Factor-β1/Bone Morphogenetic Protein 4 (TGF-β1/BMP4) Response Element within the Mouse Tissue Transglutaminase Gene Promoter. J. Biol. Chem. 1998, 273, 12798–12806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fields, G.B. Interstitial Collagen Catabolism. J. Biol. Chem. 2013, 288, 8785–8793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satpathy, M.; Shao, M.; Emerson, R.; Donner, D.B.; Matei, D. Tissue Transglutaminase Regulates Matrix Metalloproteinase-2 in Ovarian Cancer by Modulating cAMP-response Element-binding Protein Activity. J. Biol. Chem. 2009, 284, 15390–15399. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Liu, C.; Sun, Z.; Guo, X.; Zhang, Y.; Liu, M.; Li, P. CCN5 attenuates profibrotic phenotypes of fibroblasts through the Smad6-CCN2 pathway: Potential role in epidural fibrosis. Int. J. Mol. Med. 2015, 36, 123–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinnell, S.R. Regulation of collagen biosynthesis by ascorbic acid: A review. Yale J. Boil. Med. 1985, 58, 553–559. [Google Scholar]
- Macnaul, K.L.; Chartrain, N.; Lark, M.; Tocci, M.J.; Hutchinson, N.I. Discoordinate expression of stromelysin, collagenase, and tissue inhibitor of metalloproteinases-1 in rheumatoid human synovial fibroblasts. Synergistic effects of interleukin-1 and tumor necrosis factor-alpha on stromelysin expression. J. Biol. Chem. 1990, 265, 17238–17245. [Google Scholar] [CrossRef]
- Mao, J.R.; Taylor, G.; Dean, W.B.; Wagner, D.R.; Afzal, V.; Lotz, J.C.; Rubin, E.M.; Bristow, J. Tenascin-X deficiency mimics Ehlers-Danlos syndrome in mice through alteration of collagen deposition. Nat. Genet. 2002, 30, 421–425. [Google Scholar] [CrossRef]
- Liot, S.; Aubert, A.; Hervieu, V.; El Kholti, N.; Schalkwijk, J.; Verrier, B.; Valcourt, U.; Lambert, E. Loss of Tenascin-X expression during tumor progression: A new pan-cancer marker. Matrix Biol. Plus 2020, 6, 100021. [Google Scholar] [CrossRef]
- Lee, C.S.; Park, H.H. Structural aspects of transglutaminase 2: Functional, structural, and regulatory diversity. Apoptosis 2017, 22, 1057–1068. [Google Scholar] [CrossRef]
- Lim, S.; Sau, P.; Cooper, L.; McPhaden, A.; Mackenzie, K. The Incidence of Premalignant and Malignant Disease in Reinke’s Edema. Otolaryngol. Neck Surg. 2014, 150, 434–436. [Google Scholar] [CrossRef]
- Hammarstedt, A.; Hedjazifar, S.; Jenndahl, L.; Gogg, S.; Grünberg, J.; Gustafson, B.; Klimcakova, E.; Stich, V.; Langin, D.; Laakso, M.; et al. WISP2 regulates preadipocyte commitment and PPAR activation by BMP4. Proc. Natl. Acad. Sci. USA 2013, 110, 2563–2568. [Google Scholar] [CrossRef] [Green Version]
- Jeong, D.; Lee, M.-A.; Li, Y.; Yang, D.K.; Kho, C.; Oh, J.G.; Hong, G.; Lee, A.; Song, M.H.; LaRocca, T.J.; et al. Matricellular Protein CCN5 Reverses Established Cardiac Fibrosis. J. Am. Coll. Cardiol. 2016, 67, 1556–1568. [Google Scholar] [CrossRef]
- Díaz-Flores, L.; Gutiérrez, R.; García, M.D.P.; Álvarez-Argüelles, H.; López-Campos, D. CD34-positive fibroblasts in Reinke’s edema. Laryngoscope 2013, 124, E73–E80. [Google Scholar] [CrossRef] [PubMed]
- Sudo, S.; Avsian-Kretchmer, O.; Wang, L.S.; Hsueh, A.J.W. Protein Related to DAN and Cerberus Is a Bone Morphogenetic Protein Antagonist That Participates in Ovarian Paracrine Regulation. J. Biol. Chem. 2004, 279, 23134–23141. [Google Scholar] [CrossRef] [Green Version]
- Sanders, L.N.; Schoenhard, J.A.; Saleh, M.; Mukherjee, A.; Ryzhov, S.; McMaster, W.G.; Nolan, K.; Gumina, R.J.; Thompson, T.; Magnuson, M.A.; et al. BMP Antagonist Gremlin 2 Limits Inflammation After Myocardial Infarction. Circ. Res. 2016, 119, 434–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotlinowski, J.; Jozkowicz, A. PPAR Gamma and Angiogenesis: Endothelial Cells Perspective. J. Diabetes Res. 2016, 2016, 8492353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakahara, H.; Song, J.; Sugimoto, M.; Hagihara, K.; Kishimoto, T.; Yoshizaki, K.; Nishimoto, N. Anti-interleukin-6 receptor antibody therapy reduces vascular endothelial growth factor production in rheumatoid arthritis. Arthritis Rheum. 2003, 48, 1521–1529. [Google Scholar] [CrossRef] [PubMed]
- Boraschi, D.; Italiani, P.; Weil, S.; Martin, M.U. The family of the interleukin-1 receptors. Immunol. Rev. 2018, 281, 197–232. [Google Scholar] [CrossRef] [PubMed]
- Cahill, C.M.; Rogers, J.T. Interleukin (IL) 1β Induction of IL-6 Is Mediated by a Novel Phosphatidylinositol 3-Kinase-dependent AKT/IκB Kinase α Pathway Targeting Activator Protein-1. J. Biol. Chem. 2008, 283, 25900–25912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose-John, S.; Heinrich, P.C. Soluble receptors for cytokines and growth factors: Generation and biological function. Biochem. J. 1994, 300, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Ernst, M. Acquiring signalling specificity from the cytokine receptor gp130. Trends Genet. 2004, 20, 23–32. [Google Scholar] [CrossRef] [PubMed]
- He, Y.-W.; Li, H.; Zhang, J.; Hsu, C.-L.; Lin, E.; Zhang, N.; Guo, J.; Forbush, K.A.; Bevan, M.J. The extracellular matrix protein mindin is a pattern-recognition molecule for microbial pathogens. Nat. Immunol. 2003, 5, 88–97. [Google Scholar] [CrossRef]
- Zhang, Y.-L.; Li, Q.; Yang, X.-M.; Fang, F.; Li, J.; Wang, Y.-H.; Yang, Q.; Zhu, L.; Nie, H.-Z.; Zhang, X.-L.; et al. SPON2 Promotes M1-like Macrophage Recruitment and Inhibits Hepatocellular Carcinoma Metastasis by Distinct Integrin–Rho GTPase–Hippo Pathways. Cancer Res. 2018, 78, 2305–2317. [Google Scholar] [CrossRef] [Green Version]
- Artico, M.; Bronzetti, E.; Ionta, B.; Bruno, M.; Greco, A.; Ruoppolo, G.; De Virgilio, A.; Longo, L.; de Vincentiis, M. Reinke’s Edema: Investigations on the role of MIB-1 and hepatocyte growth factor. Eur. J. Histochem. 2010, 54, e30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalhal, S.; Ribeiro, S.A.; Arocena, M.; Kasciukovic, T.; Temme, A.; Koehler, K.; Huebner, A.; Griffis, E.R. The nucleoporin ALADIN regulates Aurora A localization to ensure robust mitotic spindle formation. Mol. Biol. Cell 2015, 26, 3424–3438. [Google Scholar] [CrossRef] [Green Version]
- Machida, Y.J.; Machida, Y.; Chen, Y.; Gurtan, A.M.; Kupfer, G.M.; D’Andrea, A.D.; Dutta, A. UBE2T Is the E2 in the Fanconi Anemia Pathway and Undergoes Negative Autoregulation. Mol. Cell 2006, 23, 589–596. [Google Scholar] [CrossRef]
- Roworth, A.P.; Ghari, F.; La Thangue, N.B. To live or let die—Complexity within the E2F1 pathway. Mol. Cell. Oncol. 2014, 2, e970480. [Google Scholar] [CrossRef] [PubMed]
- Mei, Q.; Huang, J.; Chen, W.; Tang, J.; Xu, C.; Yu, Q.; Cheng, Y.; Ma, L.; Yu, X.; Li, S. Regulation of DNA replication-coupled histone gene expression. Oncotarget 2017, 8, 95005–95022. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Gene Name | Company | Product Number | Sequence Primer 1 | Sequence Primer 2 |
|---|---|---|---|---|
| CCN5/WISP2 | Bio-Rad | qHsaCED0042203 | ||
| GREM2 | Bio-Rad | qHsaCED0036866 | ||
| TGM2 | Bio-Rad | qHsaCID0007428 | ||
| AQP1 | Bio-Rad | qHsaCED0005827 | ||
| SPON2 | Bio-Rad | qHsaCED0005641 | ||
| TNXB | Bio-Rad | qHsaCED0038143 | ||
| IL1R1 | Bio-Rad | qHsaCID0010015 | ||
| IL6R | Bio-Rad | qHsaCED0045907 | ||
| STAT3 | Bio-Rad | qHsaCID0010912 | ||
| SOCS3 | Bio-Rad | qHsaCED0003543 | ||
| AURKA | Bio-Rad | qHsaCID0022123 | ||
| UBE2T | Bio-Rad | qHsaCID0011482 | ||
| E2F1 | Bio-Rad | qHsaCID0016226 | ||
| CSGALNACT2 | Bio-Rad | qHsaCID0016295 | ||
| GLB1 | Bio-Rad | qHsaCID0012007 | ||
| GALNS | Bio-Rad | qHsaCID0007640 | ||
| COL1A1 | Bio-Rad | qHsaCED0002181 | ||
| TGFB1 | Microsynth | TACCTGAACCCGTGTTGCTC | GCTGAGGTATCGCCAGGAAT | |
| MMP1 | Microsynth | CACGCCAGATTTGCCAAGAG | GTTGTCCCGATGATCTCCCC | |
| MMP2 | Microsynth | ACCAAGAACTTCCGTCTGTCC | ATGTCAGGAGAGGCCCCATA | |
| MMP14 | Microsynth | CGGAGAATTTTGTGCTGCCC | AACCCTGACTCACCCCCATA | |
| COL1A2 | Microsynth | ACCACAGGGTGTTCAAGGTG | CAGGACCAGGGAGACCAAAC | |
| VWF | Microsynth | CTCATCGCAGCAAAAGGAGC | ATGCTCATGCACTCCAGGTC | |
| PECAM1 | Microsynth | CGTCGAATACCAGTGTGTTGC | TCATCCACCGGGGCTATCA | |
| NOS3 | Microsynth | CGAGTGAAGGCGACAATCCT | ACAGGACCCGGGGATCAAAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grill, M.; Lazzeri, I.; Kirsch, A.; Steurer, N.; Grossmann, T.; Karbiener, M.; Heitzer, E.; Gugatschka, M. Vocal Fold Fibroblasts in Reinke’s Edema Show Alterations Involved in Extracellular Matrix Production, Cytokine Response and Cell Cycle Control. Biomedicines 2021, 9, 735. https://doi.org/10.3390/biomedicines9070735
Grill M, Lazzeri I, Kirsch A, Steurer N, Grossmann T, Karbiener M, Heitzer E, Gugatschka M. Vocal Fold Fibroblasts in Reinke’s Edema Show Alterations Involved in Extracellular Matrix Production, Cytokine Response and Cell Cycle Control. Biomedicines. 2021; 9(7):735. https://doi.org/10.3390/biomedicines9070735
Chicago/Turabian StyleGrill, Magdalena, Isaac Lazzeri, Andrijana Kirsch, Nina Steurer, Tanja Grossmann, Michael Karbiener, Ellen Heitzer, and Markus Gugatschka. 2021. "Vocal Fold Fibroblasts in Reinke’s Edema Show Alterations Involved in Extracellular Matrix Production, Cytokine Response and Cell Cycle Control" Biomedicines 9, no. 7: 735. https://doi.org/10.3390/biomedicines9070735






