Structure and Dynamics of Zika Virus Protease and Its Insights into Inhibitor Design
Abstract
:1. Introduction
2. Protease Structure and Dynamics
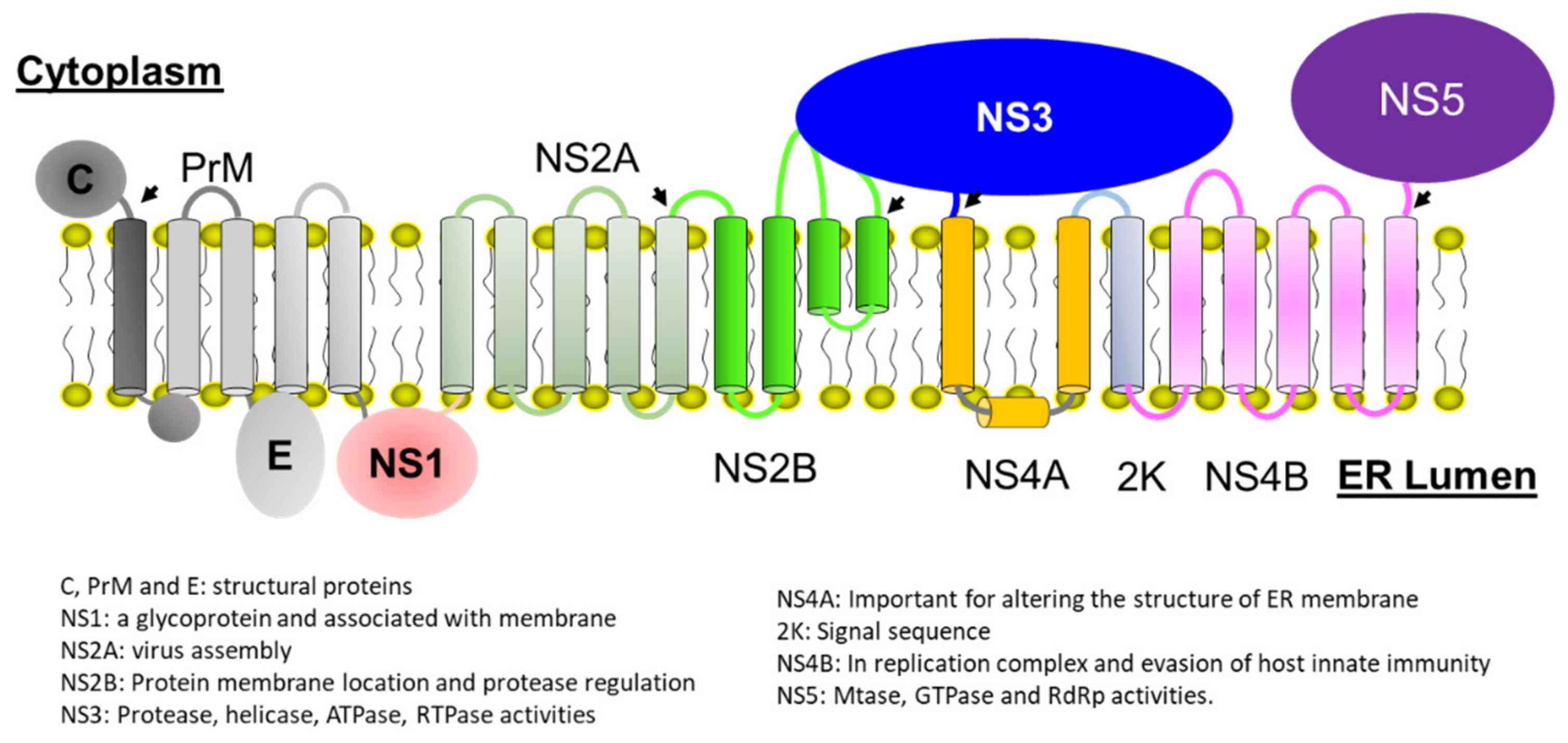
2.1. Structure of NS2B-NS3 Protease
2.2. Protease Druggability
2.3. Protease Dynamics
3. Protease Inhibitors
3.1. Peptidic Inhibitors
3.2. Small-Molecule Inhibitors
4. Strategies in Inhibitor Design
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Ndeffo-Mbah, M.L.; Parpia, A.S.; Galvani, A.P. Mitigating Prenatal Zika Virus Infection in the Americas. Ann. Intern. Med. 2016, 165, 551–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, L.R.; Jamieson, D.J.; Powers, A.M.; Honein, M.A. Zika Virus. N. Engl. J. Med. 2016, 374, 1552–1563. [Google Scholar] [CrossRef]
- Broutet, N.; Krauer, F.; Riesen, M.; Khalakdina, A.; Almiron, M.; Aldighieri, S.; Espinal, M.; Low, N.; Dye, C. Zika Virus as a Cause of Neurologic Disorders. N. Engl. J. Med. 2016, 374, 1506–1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brasil, P.; Sequeira, P.C.; Freitas, A.D.; Zogbi, H.E.; Calvet, G.A.; de Souza, R.V.; Siqueira, A.M.; de Mendonca, M.C.; Nogueira, R.M.; de Filippis, A.M.; et al. Guillain-Barre syndrome associated with Zika virus infection. Lancet 2016, 387, 1482. [Google Scholar] [CrossRef] [Green Version]
- Calvet, G.; Aguiar, R.S.; Melo, A.S.; Sampaio, S.A.; de Filippis, I.; Fabri, A.; Araujo, E.S.; de Sequeira, P.C.; de Mendonca, M.C.; de Oliveira, L.; et al. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: A case study. Lancet Infect. Dis. 2016. [Google Scholar] [CrossRef] [Green Version]
- Darko, R.; Mashburn, J.L. Zika Virus Disease: Case Report and Review of Literature. Pediatric Emerg. Care 2016, 32, 705–709. [Google Scholar] [CrossRef]
- Xie, X.; Zou, J.; Shan, C.; Yang, Y.; Kum, D.B.; Dallmeier, K.; Neyts, J.; Shi, P.Y. Zika Virus Replicons for Drug Discovery. EBioMedicine 2016, 12, 156–160. [Google Scholar] [CrossRef] [Green Version]
- Hill, M.E.; Yildiz, M.; Hardy, J.A. Cysteine Disulfide Traps Reveal Distinct Conformational Ensembles in Dengue Virus NS2B-NS3 Protease. Biochemistry 2019, 58, 776–787. [Google Scholar] [CrossRef] [PubMed]
- Karwal, P.; Vats, I.D.; Sinha, N.; Singhal, A.; Sehgal, T.; Kumari, P. Therapeutic Applications of Peptides against Zika Virus: A Review. Curr. Med. Chem. 2020, 27, 3906–3923. [Google Scholar] [CrossRef]
- Nitsche, C. Proteases from dengue, West Nile and Zika viruses as drug targets. Biophys. Rev. 2019, 11, 157–165. [Google Scholar] [CrossRef]
- Baronti, C.; Piorkowski, G.; Charrel, R.N.; Boubis, L.; Leparc-Goffart, I.; de Lamballerie, X. Complete coding sequence of zika virus from a French polynesia outbreak in 2013. Genome Announc. 2014, 2, e00500-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitatpattana, N.; Chaiyo, K.; Rajakam, S.; Poolam, K.; Chansiprasert, K.; Pesirikan, N.; Buree, S.; Rodpai, E.; Yoksan, S. Complete Genome Sequence of a Zika Virus Strain Isolated from the Serum of an Infected Patient in Thailand in 2006. Genome Announc. 2018, 6, e00121-18. [Google Scholar] [CrossRef] [Green Version]
- Sirohi, D.; Chen, Z.; Sun, L.; Klose, T.; Pierson, T.C.; Rossmann, M.G.; Kuhn, R.J. The 3.8 A resolution cryo-EM structure of Zika virus. Science 2016, 352, 467–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, R.J.; Zhang, W.; Rossmann, M.G.; Pletnev, S.V.; Corver, J.; Lenches, E.; Jones, C.T.; Mukhopadhyay, S.; Chipman, P.R.; Strauss, E.G.; et al. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell 2002, 108, 717–725. [Google Scholar] [CrossRef] [Green Version]
- Heng, C.; Huang, H.D.; Shiu, S.Y.; Chen, W.J.; Tsai, M.H.; Huang, S.H.; Wan, L.; Lin, Y.J. Functional determinants of NS2B for activation of Japanese encephalitis virus NS3 protease. Virus Res. 2007, 127, 88–94. [Google Scholar] [CrossRef]
- Cui, T.; Sugrue, R.J.; Xu, Q.; Lee, A.K.; Chan, Y.C.; Fu, J. Recombinant dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology 1998, 246, 409–417. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Lee le, T.; Wang, Q.Y.; Xie, X.; Lu, S.; Yau, Y.H.; Yuan, Z.; Geifman Shochat, S.; Kang, C.; Lescar, J.; et al. Mapping the Interactions between the NS4B and NS3 proteins of dengue virus. J. Virol. 2015, 89, 3471–3483. [Google Scholar] [CrossRef] [Green Version]
- Sampath, A.; Xu, T.; Chao, A.; Luo, D.; Lescar, J.; Vasudevan, S.G. Structure-based mutational analysis of the NS3 helicase from dengue virus. J. Virol. 2006, 80, 6686–6690. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Chang, D.C.; Hua, M.H.; Lim, S.P.; Chionh, Y.H.; Hia, F.; Lee, Y.H.; Kukkaro, P.; Lok, S.M.; Dedon, P.C.; et al. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 2012, 8, e1002642. [Google Scholar] [CrossRef] [PubMed]
- Zou, G.; Chen, Y.L.; Dong, H.; Lim, C.C.; Yap, L.J.; Yau, Y.H.; Shochat, S.G.; Lescar, J.; Shi, P.Y. Functional analysis of two cavities in flavivirus NS5 polymerase. J. Biol. Chem. 2011, 286, 14362–14372. [Google Scholar] [CrossRef] [Green Version]
- Kroschewski, H.; Lim, S.P.; Butcher, R.E.; Yap, T.L.; Lescar, J.; Wright, P.J.; Vasudevan, S.G.; Davidson, A.D. Mutagenesis of the dengue virus type 2 NS5 methyltransferase domain. J. Biol. Chem. 2008, 283, 19410–19421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawlinson, S.M.; Pryor, M.J.; Wright, P.J.; Jans, D.A. Dengue virus RNA polymerase NS5: A potential therapeutic target? Curr. Drug Targets 2006, 7, 1623–1638. [Google Scholar] [CrossRef]
- Hung, Y.F.; Schwarten, M.; Hoffmann, S.; Willbold, D.; Sklan, E.H.; Koenig, B. Amino Terminal Region of Dengue Virus NS4A Cytosolic Domain Binds to Highly Curved Liposomes. Viruses 2015, 7, 4119–4130. [Google Scholar] [CrossRef] [Green Version]
- Mackenzie, J.M.; Khromykh, A.A.; Jones, M.K.; Westaway, E.G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 1998, 245, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Ye, H.Q.; Deng, C.L.; Liu, S.Q.; Zhang, H.L.; Shang, B.D.; Shi, P.Y.; Yuan, Z.M.; Zhang, B. Genetic interaction between NS4A and NS4B for replication of Japanese encephalitis virus. J. Gen. Virol. 2015, 96, 1264–1275. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.F.; Schwarten, M.; Schunke, S.; Thiagarajan-Rosenkranz, P.; Hoffmann, S.; Sklan, E.H.; Willbold, D.; Koenig, B.W. Dengue virus NS4A cytoplasmic domain binding to liposomes is sensitive to membrane curvature. Biochim. Biophys. Acta 2015, 1848, 1119–1126. [Google Scholar] [CrossRef] [Green Version]
- Wicker, J.A.; Whiteman, M.C.; Beasley, D.W.; Davis, C.T.; McGee, C.E.; Lee, J.C.; Higgs, S.; Kinney, R.M.; Huang, C.Y.; Barrett, A.D. Mutational analysis of the West Nile virus NS4B protein. Virology 2012, 426, 22–33. [Google Scholar] [CrossRef] [Green Version]
- Moquin, S.A.; Simon, O.; Karuna, R.; Lakshminarayana, S.B.; Yokokawa, F.; Wang, F.; Saravanan, C.; Zhang, J.; Day, C.W.; Chan, K.; et al. NITD-688, a pan-serotype inhibitor of the dengue virus NS4B protein, shows favorable pharmacokinetics and efficacy in preclinical animal models. Sci. Transl. Med. 2021, 13, eabb2181. [Google Scholar] [CrossRef]
- Zou, J.; Xie, X.; Lee le, T.; Chandrasekaran, R.; Reynaud, A.; Yap, L.; Wang, Q.Y.; Dong, H.; Kang, C.; Yuan, Z.; et al. Dimerization of flavivirus NS4B protein. J. Virol. 2014, 88, 3379–3391. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.Y.; Dong, H.; Zou, B.; Karuna, R.; Wan, K.F.; Zou, J.; Susila, A.; Yip, A.; Shan, C.; Yeo, K.L.; et al. Discovery of Dengue Virus NS4B Inhibitors. J. Virol. 2015, 89, 8233–8244. [Google Scholar] [CrossRef] [Green Version]
- Munoz-Jordan, J.L.; Laurent-Rolle, M.; Ashour, J.; Martinez-Sobrido, L.; Ashok, M.; Lipkin, W.I.; Garcia-Sastre, A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 2005, 79, 8004–8013. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.; Keller, T.H.; Luo, D. Zika Virus Protease: An Antiviral Drug Target. Trends Microbiol. 2017, 25, 797–808. [Google Scholar] [CrossRef]
- Costa, S.M.; Azevedo, A.S.; Paes, M.V.; Sarges, F.S.; Freire, M.S.; Alves, A.M. DNA vaccines against dengue virus based on the ns1 gene: The influence of different signal sequences on the protein expression and its correlation to the immune response elicited in mice. Virology 2007, 358, 413–423. [Google Scholar] [CrossRef] [Green Version]
- Edeling, M.A.; Diamond, M.S.; Fremont, D.H. Structural basis of Flavivirus NS1 assembly and antibody recognition. Proc. Natl. Acad. Sci. USA 2014, 111, 4285–4290. [Google Scholar] [CrossRef] [Green Version]
- Muller, D.A.; Young, P.R. The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res. 2013, 98, 192–208. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Gayen, S.; Kang, C.; Yuan, Z.; Shi, P.Y. Membrane topology and function of dengue virus NS2A protein. J. Virol. 2013, 87, 4609–4622. [Google Scholar] [CrossRef] [Green Version]
- Vossmann, S.; Wieseler, J.; Kerber, R.; Kummerer, B.M. A basic cluster in the N terminus of yellow fever virus NS2A contributes to infectious particle production. J. Virol. 2015, 89, 4951–4965. [Google Scholar] [CrossRef] [Green Version]
- Leung, J.Y.; Pijlman, G.P.; Kondratieva, N.; Hyde, J.; Mackenzie, J.M.; Khromykh, A.A. Role of nonstructural protein NS2A in flavivirus assembly. J. Virol. 2008, 82, 4731–4741. [Google Scholar] [CrossRef] [Green Version]
- Kummerer, B.M.; Rice, C.M. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 2002, 76, 4773–4784. [Google Scholar] [CrossRef] [Green Version]
- Silva, E.M.; Conde, J.N.; Allonso, D.; Ventura, G.T.; Coelho, D.R.; Carneiro, P.H.; Silva, M.L.; Paes, M.V.; Rabelo, K.; Weissmuller, G.; et al. Dengue virus nonstructural 3 protein interacts directly with human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and reduces its glycolytic activity. Sci. Rep. 2019, 9, 2651. [Google Scholar] [CrossRef]
- Xie, X.; Zou, J.; Zhang, X.; Zhou, Y.; Routh, A.L.; Kang, C.; Popov, V.L.; Chen, X.; Wang, Q.Y.; Dong, H.; et al. Dengue NS2A Protein Orchestrates Virus Assembly. Cell Host Microbe 2019, 26, 606–622.e608. [Google Scholar] [CrossRef]
- Liang, Q.; Luo, Z.; Zeng, J.; Chen, W.; Foo, S.S.; Lee, S.A.; Ge, J.; Wang, S.; Goldman, S.A.; Zlokovic, B.V.; et al. Zika Virus NS4A and NS4B Proteins Deregulate Akt-mTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell Stem Cell 2016, 19, 663–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zmurko, J.; Neyts, J.; Dallmeier, K. Flaviviral NS4b, chameleon and jack-in-the-box roles in viral replication and pathogenesis, and a molecular target for antiviral intervention. Rev. Med. Virol. 2015, 25, 205–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchil, P.D.; Satchidanandam, V. Architecture of the Flaviviral Replication Complex: Protease, Nuclease, And Detergents Reveal Encasement within Double-Layered Membrane Compartments. J. Biol. Chem. 2003, 278, 24388–24398. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-D.; Deng, C.-L.; Ye, H.-Q.; Zhang, H.-L.; Zhang, Q.-Y.; Chen, D.-D.; Zhang, P.-T.; Shi, P.-Y.; Yuan, Z.-M.; Zhang, B. Transmembrane Domains of NS2B Contribute to both Viral RNA Replication and Particle Formation in Japanese Encephalitis Virus. J. Virol. 2016, 90, 5735–5749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitsche, C. Strategies Towards Protease Inhibitors for Emerging Flaviviruses. In Dengue and Zika: Control and Antiviral Treatment Strategies; Hilgenfeld, R., Vasudevan, S.G., Eds.; Springer Singapore: Singapore, 2018; pp. 175–186. [Google Scholar] [CrossRef]
- Lim, S.P. Dengue drug discovery: Progress, challenges and outlook. Antivir. Res. 2019, 163, 156–178. [Google Scholar] [CrossRef] [PubMed]
- Majerova, T.; Novotny, P.; Krysova, E.; Konvalinka, J. Exploiting the unique features of Zika and Dengue proteases for inhibitor design. Biochimie 2019, 166, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Huo, T.; Lin, Y.L.; Nie, S.; Wu, F.; Hua, Y.; Wu, J.; Kneubehl, A.R.; Vogt, M.B.; Rico-Hesse, R.; et al. Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease. J. Am. Chem. Soc. 2019, 141, 6832–6836. [Google Scholar] [CrossRef] [PubMed]
- Abrams, R.P.M.; Yasgar, A.; Teramoto, T.; Lee, M.-H.; Dorjsuren, D.; Eastman, R.T.; Malik, N.; Zakharov, A.V.; Li, W.; Bachani, M.; et al. Therapeutic candidates for the Zika virus identified by a high-throughput screen for Zika protease inhibitors. Proc. Natl. Acad. Sci. USA 2020, 117, 31365–31375. [Google Scholar] [CrossRef] [PubMed]
- Voss, S.; Nitsche, C. Inhibitors of the Zika virus protease NS2B-NS3. Bioorg. Med. Chem. Lett. 2020, 30, 126965. [Google Scholar] [CrossRef]
- Lima, C.S.; Mottin, M.; de Assis, L.R.; Mesquita, N.C.d.M.R.; Sousa, B.K.d.P.; Coimbra, L.D.; Santos, K.B.-d.; Zorn, K.M.; Guido, R.V.C.; Ekins, S.; et al. Flavonoids from Pterogyne nitens as Zika virus NS2B-NS3 protease inhibitors. Bioorg. Chem. 2021, 109, 104719. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Hansen, G.; Nitsche, C.; Klein, C.D.; Zhang, L.; Hilgenfeld, R. Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science 2016, 353, 503–505. [Google Scholar] [CrossRef] [Green Version]
- Akaberi, D.; Bahlstrom, A.; Chinthakindi, P.K.; Nyman, T.; Sandstrom, A.; Jarhult, J.D.; Palanisamy, N.; Lundkvist, A.; Lennerstrand, J. Targeting the NS2B-NS3 protease of tick-borne encephalitis virus with pan-flaviviral protease inhibitors. Antivir. Res. 2021, 190, 105074. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, K.; Wu, C.; Chen, C.; Hu, C.; Buzovetsky, O.; Wang, Z.; Ji, X.; Xiong, Y.; Yang, H. Mechanisms of activation and inhibition of Zika virus NS2B-NS3 protease. Cell Res. 2016, 26, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Li, Y.; Loh, Y.R.; Phoo, W.W.; Hung, A.W.; Kang, C.; Luo, D. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science 2016, 354, 1597–1600. [Google Scholar] [CrossRef]
- Phoo, W.W.; Li, Y.; Zhang, Z.; Lee, M.Y.; Loh, Y.R.; Tan, Y.B.; Ng, E.Y.; Lescar, J.; Kang, C.; Luo, D. Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat. Commun. 2016, 7, 13410. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Z.; Phoo, W.W.; Loh, Y.R.; Li, R.; Yang, H.Y.; Jansson, A.E.; Hill, J.; Keller, T.H.; Nacro, K.; et al. Structural Insights into the Inhibition of Zika Virus NS2B-NS3 Protease by a Small-Molecule Inhibitor. Structure 2018, 26, 555–564. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, Z.; Phoo, W.W.; Loh, Y.R.; Wang, W.; Liu, S.; Chen, M.W.; Hung, A.W.; Keller, T.H.; Luo, D.; et al. Structural Dynamics of Zika Virus NS2B-NS3 Protease Binding to Dipeptide Inhibitors. Structure 2017, 25, 1242–1250.e1243. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Phoo, W.W.; Loh, Y.R.; Zhang, Z.; Ng, E.Y.; Wang, W.; Keller, T.H.; Luo, D.; Kang, C. Structural characterization of the linked NS2B-NS3 protease of Zika virus. FEBS Lett. 2017, 591, 2338–2347. [Google Scholar] [CrossRef] [Green Version]
- Su, X.C.; Ozawa, K.; Qi, R.; Vasudevan, S.G.; Lim, S.P.; Otting, G. NMR analysis of the dynamic exchange of the NS2B cofactor between open and closed conformations of the West Nile virus NS2B-NS3 protease. PLoS Negl. Trop. Dis. 2009, 3, e561. [Google Scholar] [CrossRef]
- Mahawaththa, M.C.; Pearce, B.J.; Szabo, M.; Graham, B.; Klein, C.D.; Nitsche, C.; Otting, G. Solution conformations of a linked construct of the Zika virus NS2B-NS3 protease. Antivir. Res. 2017, 142, 141–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.N.; Loscha, K.V.; Nitsche, C.; Graham, B.; Otting, G. The dengue virus NS2B-NS3 protease retains the closed conformation in the complex with BPTI. FEBS Lett. 2014, 588, 2206–2211. [Google Scholar] [CrossRef] [Green Version]
- Su, X.C.; Ozawa, K.; Yagi, H.; Lim, S.P.; Wen, D.; Ekonomiuk, D.; Huang, D.; Keller, T.H.; Sonntag, S.; Caflisch, A.; et al. NMR study of complexes between low molecular mass inhibitors and the West Nile virus NS2B-NS3 protease. FEBS J. 2009, 276, 4244–4255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de la Cruz, L.; Chen, W.N.; Graham, B.; Otting, G. Binding mode of the activity-modulating C-terminal segment of NS2B to NS3 in the dengue virus NS2B-NS3 protease. FEBS J. 2014, 281, 1517–1533. [Google Scholar] [CrossRef] [PubMed]
- Erbel, P.; Schiering, N.; D’Arcy, A.; Renatus, M.; Kroemer, M.; Lim, S.P.; Yin, Z.; Keller, T.H.; Vasudevan, S.G.; Hommel, U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006, 13, 372–373. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-R.; Lai, Y.-C.; Yeh, T.-M. Dengue virus non-structural protein 1: A pathogenic factor, therapeutic target, and vaccine candidate. J. Biomed. Sci. 2018, 25, 58. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wong, Y.L.; Lee, M.Y.; Li, Q.; Wang, Q.Y.; Lescar, J.; Shi, P.Y.; Kang, C. Secondary Structure and Membrane Topology of the Full-Length Dengue Virus NS4B in Micelles. Angew. Chem. Int. Ed. Engl. 2016, 55, 12068–12072. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Xie, X.; Wang, Q.Y.; Dong, H.; Lee, M.Y.; Kang, C.; Yuan, Z.; Shi, P.Y. Characterization of dengue virus NS4A and NS4B protein interaction. J. Virol. 2015, 89, 3455–3470. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, Q.; Wong, Y.L.; Liew, L.S.; Kang, C. Membrane topology of NS2B of dengue virus revealed by NMR spectroscopy. Biochim. Biophys. Acta 2015, 1848, 2244–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Kim, Y.M.; Zou, J.; Wang, Q.Y.; Gayen, S.; Wong, Y.L.; Lee le, T.; Xie, X.; Huang, Q.; Lescar, J.; et al. Secondary structure and membrane topology of dengue virus NS4B N-terminal 125 amino acids. Biochim. Biophys. Acta 2015, 1848, 3150–3157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.M.; Xie, X.; Zou, J.; Li, S.H.; Lee, M.Y.; Dong, H.; Qin, C.F.; Kang, C.; Shi, P.Y. Determinants of Dengue Virus NS4A Protein Oligomerization. J. Virol. 2015, 89, 6171–6183. [Google Scholar] [CrossRef] [Green Version]
- Yildiz, M.; Ghosh, S.; Bell, J.A.; Sherman, W.; Hardy, J.A. Allosteric Inhibition of the NS2B-NS3 Protease from Dengue Virus. ACS Chem. Biol. 2013, 17, 76–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Z.; Lim, S.P.; Patel, S.; Patel, V.; Beer, D.; Ma, N.L.; Vasudevan, S.; Keller, T. Targeting the protease activity of Dengue virus NS3. Acta Pharmacol. Sin. 2006, 27, 251. [Google Scholar]
- Lescar, J.; Luo, D.; Xu, T.; Sampath, A.; Lim, S.P.; Canard, B.; Vasudevan, S.G. Towards the design of antiviral inhibitors against flaviviruses: The case for the multifunctional NS3 protein from Dengue virus as a target. Antivir. Res. 2008, 80, 94–101. [Google Scholar] [CrossRef]
- Shiryaev, S.A.; Kozlov, I.A.; Ratnikov, B.I.; Smith, J.W.; Lebl, M.; Strongin, A.Y. Cleavage preference distinguishes the two-component NS2B-NS3 serine proteinases of Dengue and West Nile viruses. Biochem. J. 2007, 401, 743–752. [Google Scholar] [CrossRef]
- Chappell, K.J.; Stoermer, M.J.; Fairlie, D.P.; Young, P.R. West Nile Virus NS2B/NS3 protease as an antiviral target. Curr. Med. Chem. 2008, 15, 2771–2784. [Google Scholar] [CrossRef]
- Li, H.; Zhu, L.; Hou, S.; Yang, J.; Wang, J.; Liu, J. An inhibition model of BPTI to unlinked dengue virus NS2B-NS3 protease. FEBS Lett. 2014, 588, 2794–2799. [Google Scholar] [CrossRef] [Green Version]
- Yusof, R.; Clum, S.; Wetzel, M.; Murthy, H.M.; Padmanabhan, R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J. Biol. Chem. 2000, 275, 9963–9969. [Google Scholar] [CrossRef] [Green Version]
- Leung, D.; Schroder, K.; White, H.; Fang, N.X.; Stoermer, M.J.; Abbenante, G.; Martin, J.L.; Young, P.R.; Fairlie, D.P. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J. Biol. Chem. 2001, 276, 45762–45771. [Google Scholar] [CrossRef] [Green Version]
- Clum, S.; Ebner, K.E.; Padmanabhan, R. Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J. Biol. Chem. 1997, 272, 30715–30723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aktepe, T.E.; Mackenzie, J.M. Shaping the flavivirus replication complex: It is curvaceous! Cell. Microbiol. 2018, 20, e12884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lescar, J.; Soh, S.; Lee, L.T.; Vasudevan, S.G.; Kang, C.; Lim, S.P. The Dengue Virus Replication Complex: From RNA Replication to Protein-Protein Interactions to Evasion of Innate Immunity. In Dengue and Zika: Control and Antiviral Treatment Strategies; Hilgenfeld, R., Vasudevan, S.G., Eds.; Springer Singapore: Singapore, 2018; pp. 115–129. [Google Scholar] [CrossRef]
- Ngo, A.M.; Shurtleff, M.J.; Popova, K.D.; Kulsuptrakul, J.; Weissman, J.S.; Puschnik, A.S. The ER membrane protein complex is required to ensure correct topology and stable expression of flavivirus polyproteins. eLife 2019, 8, e48469. [Google Scholar] [CrossRef] [PubMed]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.E.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and Three-Dimensional Architecture of the Dengue Virus Replication and Assembly Sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [Green Version]
- van den Elsen, K.; Quek, J.P.; Luo, D. Molecular Insights into the Flavivirus Replication Complex. Viruses 2021, 13, 956. [Google Scholar] [CrossRef]
- Pant, A.; Pasupureddy, R.; Pande, V.; Seshadri, S.; Dixit, R.; Pandey, K.C. Proteases in Mosquito Borne Diseases: New Avenues in Drug Development. Curr. Top. Med. Chem. 2017, 17, 2221–2232. [Google Scholar] [CrossRef]
- Behnam, M.A.M.; Klein, C.D.P. Conformational selection in the flaviviral NS2B-NS3 protease. Biochimie 2020, 174, 117–125. [Google Scholar] [CrossRef]
- Ng, E.Y.; Loh, Y.R.; Li, Y.; Li, Q.; Kang, C. Expression, purification of Zika virus membrane protein-NS2B in detergent micelles for NMR studies. Protein Expr. Purif. 2019, 154, 1–6. [Google Scholar] [CrossRef]
- Luo, D.; Xu, T.; Hunke, C.; Gruber, G.; Vasudevan, S.G.; Lescar, J. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 2008, 82, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Robin, G.; Chappell, K.; Stoermer, M.J.; Hu, S.H.; Young, P.R.; Fairlie, D.P.; Martin, J.L. Structure of West Nile virus NS3 protease: Ligand stabilization of the catalytic conformation. J. Mol. Biol. 2009, 385, 1568–1577. [Google Scholar] [CrossRef]
- Li, L.; Basavannacharya, C.; Chan, K.W.; Shang, L.; Vasudevan, S.G.; Yin, Z. Structure-guided Discovery of a Novel Non-peptide Inhibitor of Dengue Virus NS2B-NS3 Protease. Chem. Biol. Drug Des. 2015, 86, 255–264. [Google Scholar] [CrossRef]
- Nall, T.A.; Chappell, K.J.; Stoermer, M.J.; Fang, N.X.; Tyndall, J.D.; Young, P.R.; Fairlie, D.P. Enzymatic characterization and homology model of a catalytically active recombinant West Nile virus NS3 protease. J. Biol. Chem. 2004, 279, 48535–48542. [Google Scholar] [CrossRef] [Green Version]
- Noble, C.G.; Seh, C.C.; Chao, A.T.; Shi, P.Y. Ligand-bound structures of the dengue virus protease reveal the active conformation. J. Virol. 2012, 86, 438–446. [Google Scholar] [CrossRef] [Green Version]
- Hilgenfeld, R.; Lei, J.; Zhang, L. The Structure of the Zika Virus Protease, NS2B/NS3(pro). Adv. Exp. Med. Biol. 2018, 1062, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Shannon, A.E.; Chappell, K.J.; Stoermer, M.J.; Chow, S.Y.; Kok, W.M.; Fairlie, D.P.; Young, P.R. Simultaneous uncoupled expression and purification of the Dengue virus NS3 protease and NS2B co-factor domain. Protein Expr. Purif. 2015, 119, 124–129. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.M.; Gayen, S.; Kang, C.; Joy, J.; Huang, Q.; Chen, A.S.; Wee, J.L.; Ang, M.J.; Lim, H.A.; Hung, A.W.; et al. NMR Analysis of a Novel Enzymatically Active Unlinked Dengue NS2B-NS3 Protease Complex. J. Biol. Chem. 2013, 288, 12891–12900. [Google Scholar] [CrossRef] [Green Version]
- . Tseng, A.C.; Nerurkar, V.R.; Neupane, K.R.; Kae, H.; Kaufusi, P.H. Potential Dual Role of West Nile Virus NS2B in Orchestrating NS3 Enzymatic Activity in Viral Replication. Viruses 2021, 13, 216. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Li, X.F.; Ye, H.Q.; Deng, C.L.; Ye, Q.; Shan, C.; Shang, B.D.; Xu, L.L.; Li, S.H.; Cao, S.B.; et al. Recovery of a chemically synthesized Japanese encephalitis virus reveals two critical adaptive mutations in NS2B and NS4A. J. Gen. Virol. 2014, 95, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Fan, J.; Zhang, B.; Yuan, Z. Mutagenesis of D80-82 and G83 residues in West Nile Virus NS2B: Effects on NS2B-NS3 activity and viral replication. Virol. Sin. 2013, 28, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Quek, J.P.; Liu, S.; Zhang, Z.; Li, Y.; Ng, E.Y.; Loh, Y.R.; Hung, A.W.; Luo, D.; Kang, C. Identification and structural characterization of small molecule fragments targeting Zika virus NS2B-NS3 protease. Antivir. Res. 2020, 175, 104707. [Google Scholar] [CrossRef] [PubMed]
- de la Cruz, L.; Nguyen, T.H.; Ozawa, K.; Shin, J.; Graham, B.; Huber, T.; Otting, G. Binding of low molecular weight inhibitors promotes large conformational changes in the dengue virus NS2B-NS3 protease: Fold analysis by pseudocontact shifts. J. Am. Chem. Soc. 2011, 133, 19205–19215. [Google Scholar] [CrossRef]
- Ekonomiuk, D.; Su, X.C.; Ozawa, K.; Bodenreider, C.; Lim, S.P.; Otting, G.; Huang, D.; Caflisch, A. Flaviviral protease inhibitors identified by fragment-based library docking into a structure generated by molecular dynamics. J. Med. Chem. 2009, 52, 4860–4868. [Google Scholar] [CrossRef]
- Pilla, K.B.; Leman, J.K.; Otting, G.; Huber, T. Capturing conformational States in proteins using sparse paramagnetic NMR data. PLoS ONE 2015, 10, e0127053. [Google Scholar] [CrossRef] [PubMed]
- von Hammerstein, F.; Lauth, L.M.; Hammerschmidt, S.; Wagner, A.; Schirmeister, T.; Hellmich, U.A. Cis autocatalytic cleavage of glycine-linked Zika virus NS2B-NS3 protease constructs. FEBS Lett. 2019, 593, 2204–2213. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Ren, J.; Nocadello, S.; Rice, A.J.; Ojeda, I.; Light, S.; Minasov, G.; Vargas, J.; Nagarathnam, D.; Anderson, W.F.; et al. Identification of novel small molecule inhibitors against NS2B/NS3 serine protease from Zika virus. Antivir. Res. 2017, 139, 49–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Loh, Y.R.; Hung, A.W.; Kang, C. Characterization of molecular interactions between Zika virus protease and peptides derived from the C-terminus of NS2B. Biochem. Biophys. Res. Commun. 2018, 503, 691–696. [Google Scholar] [CrossRef]
- Kuiper, B.D.; Slater, K.; Spellmon, N.; Holcomb, J.; Medapureddy, P.; Muzzarelli, K.M.; Yang, Z.; Ovadia, R.; Amblard, F.; Kovari, I.A.; et al. Increased activity of unlinked Zika virus NS2B/NS3 protease compared to linked Zika virus protease. Biochem. Biophys. Res. Commun. 2017, 492, 668–673. [Google Scholar] [CrossRef]
- Li, Q.; Kang, C. Insights into Structures and Dynamics of Flavivirus Proteases from NMR Studies. Int. J. Mol. Sci. 2020, 21, 2527. [Google Scholar] [CrossRef] [Green Version]
- Owens, J. Determining druggability. Nat. Rev. Drug Discov. 2007, 6, 187. [Google Scholar] [CrossRef]
- Mulgaonkar, N.; Wang, H.; King, M.; Fernando, S. Druggability assessment of precursor membrane protein as a target for inhibiting the Zika virus. J. Biomol. Struct. Dyn. 2020, 1–17. [Google Scholar] [CrossRef]
- Fauman, E.B.; Rai, B.K.; Huang, E.S. Structure-based druggability assessment—identifying suitable targets for small molecule therapeutics. Curr. Opin. Chem. Biol. 2011, 15, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Wehrhan, L.; Hillisch, A.; Mundt, S.; Tersteegen, A.; Meier, K. Druggability Assessment for Selected Serine Proteases in a Pharmaceutical Industry Setting. ChemMedChem 2020, 15, 2010–2018. [Google Scholar] [CrossRef] [PubMed]
- Volkamer, A.; Kuhn, D.; Rippmann, F.; Rarey, M. DoGSiteScorer: A web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics 2012, 28, 2074–2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, C.; Gayen, S.; Wang, W.; Severin, R.; Chen, A.S.; Lim, H.A.; Chia, C.S.; Schuller, A.; Doan, D.N.; Poulsen, A.; et al. Exploring the binding of peptidic West Nile virus NS2B-NS3 protease inhibitors by NMR. Antivir. Res. 2013, 97, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, A.; Kang, C.; Keller, T.H. Drug design for flavivirus proteases: What are we missing? Curr. Pharm. Des. 2014, 20, 3422–3427. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.; Dang, M.; Roy, A.; Kang, J.; Song, J. Curcumin Allosterically Inhibits the Dengue NS2B-NS3 Protease by Disrupting Its Active Conformation. ACS Omega 2020, 5, 25677–25686. [Google Scholar] [CrossRef]
- Nitsche, C.; Passioura, T.; Varava, P.; Mahawaththa, M.C.; Leuthold, M.M.; Klein, C.D.; Suga, H.; Otting, G. De Novo Discovery of Nonstandard Macrocyclic Peptides as Noncompetitive Inhibitors of the Zika Virus NS2B-NS3 Protease. ACS Med. Chem. Lett. 2019, 10, 168–174. [Google Scholar] [CrossRef]
- Othman, R.; Kiat, T.S.; Khalid, N.; Yusof, R.; Newhouse, E.I.; Newhouse, J.S.; Alam, M.; Rahman, N.A. Docking of noncompetitive inhibitors into dengue virus type 2 protease: Understanding the interactions with allosteric binding sites. J. Chem. Inf. Model. 2008, 48, 1582–1591. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Li, Q.; Joy, J.; Chen, A.S.; Ruiz-Carrillo, D.; Hill, J.; Lescar, J.; Kang, C. Lyso-myristoyl phosphatidylcholine micelles sustain the activity of Dengue non-structural (NS) protein 3 protease domain fused with the full-length NS2B. Protein Expr. Purif. 2013, 92, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Chen, A.S.; Li, Q.; Kang, C. Expression, purification, and initial structural characterization of nonstructural protein 2B, an integral membrane protein of Dengue-2 virus, in detergent micelles. Protein Expr. Purif. 2011, 80, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.H.; Caffarena, E.R.; Ferreira, R.S. pH and non-covalent ligand binding modulate Zika virus NS2B/NS3 protease binding site residues: Discoveries from MD and constant pH MD simulations. J. Biomol. Struct. Dyn. 2021, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.N.; Nitsche, C.; Pilla, K.B.; Graham, B.; Huber, T.; Klein, C.D.; Otting, G. Sensitive NMR Approach for Determining the Binding Mode of Tightly Binding Ligand Molecules to Protein Targets. J. Am. Chem. Soc. 2016, 138, 4539–4546. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.; Lee, H.; Kotak, A.; Johnson, M.E. MD simulations reveal alternate conformations of the oxyanion hole in the Zika virus NS2B/NS3 protease. Proteins 2020, 88, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Nutho, B.; Rungrotmongkol, T. Binding recognition of substrates in NS2B/NS3 serine protease of Zika virus revealed by molecular dynamics simulations. J. Mol. Graph. Model. 2019, 92, 227–235. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, L.R.; Wu, H.; Nebo, L.; Fernandes, J.B.; da Silva, M.F.; Kiefer, W.; Kanitz, M.; Bodem, J.; Diederich, W.E.; Schirmeister, T.; et al. Flavonoids as noncompetitive inhibitors of Dengue virus NS2B-NS3 protease: Inhibition kinetics and docking studies. Bioorg. Med. Chem. 2015, 23, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Campos, D.M.O.; Bezerra, K.S.; Esmaile, S.C.; Fulco, U.L.; Albuquerque, E.L.; Oliveira, J.I.N. Intermolecular interactions of cn-716 and acyl-KR-aldehyde dipeptide inhibitors against Zika virus. Phys. Chem. Chem. Phys. 2020, 22, 15683–15695. [Google Scholar] [CrossRef]
- Brecher, M.; Zhang, J.; Li, H. The flavivirus protease as a target for drug discovery. Virol. Sin. 2013, 28, 326–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, H.J.; Kim, M.H.; Lee, J.Y.; Hwang, I.; Yoon, G.Y.; Kim, H.S.; Kwon, Y.C.; Ahn, D.G.; Kim, K.D.; Kim, B.T.; et al. Structure-Based Virtual Screening: Identification of a Novel NS2B-NS3 Protease Inhibitor with Potent Antiviral Activity against Zika and Dengue Viruses. Microorganisms 2021, 9, 545. [Google Scholar] [CrossRef] [PubMed]
- Sisakht, M.; Mahmoodzadeh, A.; Darabian, M. Plant-derived chemicals as potential inhibitors of SARS-CoV-2 main protease (6LU7), a virtual screening study. Phytother. Res. 2021, 35, 3262–3274. [Google Scholar] [CrossRef] [PubMed]
- Pathak, N.; Kuo, Y.P.; Chang, T.Y.; Huang, C.T.; Hung, H.C.; Hsu, J.T.; Yu, G.Y.; Yang, J.M. Zika Virus NS3 Protease Pharmacophore Anchor Model and Drug Discovery. Sci. Rep. 2020, 10, 8929. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Selvaraj, C.; Aarthy, M.; Kumar, P.; Kumar, A.; Singh, S.K.; Giri, R. Investigating into the molecular interactions of flavonoids targeting NS2B-NS3 protease from ZIKA virus through in-silico approaches. J. Biomol. Struct. Dyn. 2021, 39, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Behnam, M.A.M.; Nitsche, C.; Boldescu, V.; Klein, C.D. The Medicinal Chemistry of Dengue Virus. J. Med. Chem. 2016, 59, 5622–5649. [Google Scholar] [CrossRef] [Green Version]
- Boldescu, V.; Behnam, M.A.M.; Vasilakis, N.; Klein, C.D. Broad-spectrum agents for flaviviral infections: Dengue, Zika and beyond. Nat. Rev. Drug Discov. 2017, 16, 565–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.P.; Shi, P.Y. West Nile virus drug discovery. Viruses 2013, 5, 2977–3006. [Google Scholar] [CrossRef] [Green Version]
- Nitsche, C.; Holloway, S.; Schirmeister, T.; Klein, C.D. Biochemistry and Medicinal Chemistry of the Dengue Virus Protease. Chem. Rev. 2014. [Google Scholar] [CrossRef] [PubMed]
- Timiri, A.K.; Sinha, B.N.; Jayaprakash, V. Progress and prospects on DENV protease inhibitors. Eur. J. Med. Chem. 2016, 117, 125–143. [Google Scholar] [CrossRef] [PubMed]
- Rassias, G.; Zogali, V.; Swarbrick, C.M.D.; Ki Chan, K.W.; Chan, S.A.; Gwee, C.P.; Wang, S.; Kaplanai, E.; Canko, A.; Kiousis, D.; et al. Cell-active carbazole derivatives as inhibitors of the zika virus protease. Eur. J. Med. Chem. 2019, 180, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Yao, Y.; Wu, F.; Wu, X.; Zhao, J.; Hua, Y.; Wu, J.; Huo, T.; Lin, Y.-L.; Kneubehl, A.R.; et al. Synthesis, Structure–Activity Relationships, and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B–NS3 Protease. J. Med. Chem. 2021, 64, 2777–2800. [Google Scholar] [CrossRef]
- Millies, B.; von Hammerstein, F.; Gellert, A.; Hammerschmidt, S.; Barthels, F.; Goppel, U.; Immerheiser, M.; Elgner, F.; Jung, N.; Basic, M.; et al. Proline-Based Allosteric Inhibitors of Zika and Dengue Virus NS2B/NS3 Proteases. J. Med. Chem. 2019, 62, 11359–11382. [Google Scholar] [CrossRef] [PubMed]
- Braun, N.J.; Quek, J.P.; Huber, S.; Kouretova, J.; Rogge, D.; Lang-Henkel, H.; Cheong, E.Z.K.; Chew, B.L.A.; Heine, A.; Luo, D.; et al. Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses. ChemMedChem 2020, 15, 1439–1452. [Google Scholar] [CrossRef]
- Patil, N.A.; Quek, J.P.; Schroeder, B.; Morewood, R.; Rademann, J.; Luo, D.; Nitsche, C. 2-Cyanoisonicotinamide Conjugation: A Facile Approach to Generate Potent Peptide Inhibitors of the Zika Virus Protease. ACS Med. Chem. Lett. 2021, 12, 732–737. [Google Scholar] [CrossRef] [PubMed]
- da Silva-Junior, E.F.; de Araujo-Junior, J.X. Peptide derivatives as inhibitors of NS2B-NS3 protease from Dengue, West Nile, and Zika flaviviruses. Bioorg. Med. Chem. 2019, 27, 3963–3978. [Google Scholar] [CrossRef]
- Knox, J.E.; Ma, N.L.; Yin, Z.; Patel, S.J.; Wang, W.L.; Chan, W.L.; Rao, K.R.R.; Wang, G.; Ngew, X.; Patel, V.; et al. Peptide inhibitors of West Nile NS3 protease: SAR study of tetrapeptide aldehyde inhibitors. J. Med. Chem. 2006, 49, 6585–6590. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Patel, S.J.; Wang, W.L.; Wang, G.; Chan, W.L.; Rao, K.R.; Alam, J.; Jeyaraj, D.A.; Ngew, X.; Patel, V.; et al. Peptide inhibitors of Dengue virus NS3 protease. Part 1: Warhead. Bioorg. Med. Chem. Lett. 2006, 16, 36–39. [Google Scholar] [CrossRef]
- Schuller, A.; Yin, Z.; Brian Chia, C.S.; Doan, D.N.; Kim, H.K.; Shang, L.; Loh, T.P.; Hill, J.; Vasudevan, S.G. Tripeptide inhibitors of dengue and West Nile virus NS2B-NS3 protease. Antivir. Res. 2011, 92, 96–101. [Google Scholar] [CrossRef]
- Mushtaq, M.; Naz, S.; Parang, K.; Ul-Haq, Z. Exploiting Dengue Virus Protease as a Therapeutic Target; Current Status, Challenges and Future Avenues. Curr. Med. Chem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Drazic, T.; Kopf, S.; Corridan, J.; Leuthold, M.M.; Bertosa, B.; Klein, C.D. Peptide-beta-lactam Inhibitors of Dengue and West Nile Virus NS2B-NS3 Protease Display Two Distinct Binding Modes. J. Med. Chem. 2020, 63, 140–156. [Google Scholar] [CrossRef]
- Li, Z.; Xu, J.; Lang, Y.; Fan, X.; Kuo, L.; D'Brant, L.; Hu, S.; Samrat, S.K.; Trudeau, N.; Tharappel, A.M.; et al. JMX0207, a Niclosamide Derivative with Improved Pharmacokinetics, Suppresses Zika Virus Infection Both In Vitro and In Vivo. ACS Infect. Dis. 2020, 6, 2616–2628. [Google Scholar] [CrossRef]
- Baltina, L.A.; Lai, H.C.; Liu, Y.C.; Huang, S.H.; Hour, M.J.; Baltina, L.A.; Nugumanov, T.R.; Borisevich, S.S.; Khalilov, L.M.; Petrova, S.F.; et al. Glycyrrhetinic acid derivatives as Zika virus inhibitors: Synthesis and antiviral activity in vitro. Bioorg. Med. Chem. 2021, 41, 116204. [Google Scholar] [CrossRef]
- Koh-Stenta, X.; Joy, J.; Wang, S.F.; Kwek, P.Z.; Wee, J.L.; Wan, K.F.; Gayen, S.; Chen, A.S.; Kang, C.; Lee, M.A.; et al. Identification of covalent active site inhibitors of dengue virus protease. Drug Des. Devel. Ther. 2015, 9, 6389–6399. [Google Scholar] [CrossRef] [Green Version]
- Mukhametov, A.; Newhouse, E.I.; Aziz, N.A.; Saito, J.A.; Alam, M. Allosteric pocket of the dengue virus (serotype 2) NS2B/NS3 protease: In silico ligand screening and molecular dynamics studies of inhibition. J. Mol. Graph. Model. 2014, 52, 103–113. [Google Scholar] [CrossRef]
- Brecher, M.; Li, Z.; Liu, B.; Zhang, J.; Koetzner, C.A.; Alifarag, A.; Jones, S.A.; Lin, Q.; Kramer, L.D.; Li, H. A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PLoS Pathog. 2017, 13, e1006411. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, H.; Alzahrani, F.A.; Hassan, M.A.; Alghamdi, A.; Abdulaal, W.H.; Bakhrebah, M.A.; Zamzami, M.A.; Helmi, N.; Bokhari, F.F.; Zeyadi, M.; et al. Zika Virus Targeting by Screening Inhibitors against NS2B/NS3 Protease. Biomed. Res. Int. 2019, 2019, 3947245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akaberi, D.; Chinthakindi, P.K.; Bahlstrom, A.; Palanisamy, N.; Sandstrom, A.; Lundkvist, A.; Lennerstrand, J. Identification of a C2-symmetric diol based human immunodeficiency virus protease inhibitor targeting Zika virus NS2B-NS3 protease. J. Biomol. Struct. Dyn. 2020, 38, 5526–5536. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.R.S.; Nunes, D.A.F.; Lima, W.G.; Davyt, D.; Santos, L.L.; Taranto, A.G.; MS Ferreira, J. Identification of Zika Virus NS2B-NS3 Protease Inhibitors by Structure-Based Virtual Screening and Drug Repurposing Approaches. J. Chem. Inf. Model. 2020, 60, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Sinigaglia, A.; Riccetti, S.; Trevisan, M.; Barzon, L. In silico approaches to Zika virus drug discovery. Expert Opin. Drug Discov. 2018, 13, 825–835. [Google Scholar] [CrossRef]
- Hasan, S.S.; Sevvana, M.; Kuhn, R.J.; Rossmann, M.G. Structural biology of Zika virus and other flaviviruses. Nat. Struct. Mol. Biol. 2018, 25, 13–20. [Google Scholar] [CrossRef]
- Yuan, S.; Chan, J.F.; den-Haan, H.; Chik, K.K.; Zhang, A.J.; Chan, C.C.; Poon, V.K.; Yip, C.C.; Mak, W.W.; Zhu, Z.; et al. Structure-based discovery of clinically approved drugs as Zika virus NS2B-NS3 protease inhibitors that potently inhibit Zika virus infection in vitro and in vivo. Antivir. Res. 2017, 145, 33–43. [Google Scholar] [CrossRef]
- Chu, J.J.; Lee, R.C.; Ang, M.J.; Wang, W.L.; Lim, H.A.; Wee, J.L.; Joy, J.; Hill, J.; Brian Chia, C.S. Antiviral activities of 15 dengue NS2B-NS3 protease inhibitors using a human cell-based viral quantification assay. Antivir. Res. 2015, 118, 68–74. [Google Scholar] [CrossRef]
- Shiryaev, S.A.; Farhy, C.; Pinto, A.; Huang, C.-T.; Simonetti, N.; Ngono, A.E.; Dewing, A.; Shresta, S.; Pinkerton, A.B.; Cieplak, P.; et al. Characterization of the Zika virus two-component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antivir. Res. 2017, 143, 218–229. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Kang, C. Structure and Dynamics of Zika Virus Protease and Its Insights into Inhibitor Design. Biomedicines 2021, 9, 1044. https://doi.org/10.3390/biomedicines9081044
Li Q, Kang C. Structure and Dynamics of Zika Virus Protease and Its Insights into Inhibitor Design. Biomedicines. 2021; 9(8):1044. https://doi.org/10.3390/biomedicines9081044
Chicago/Turabian StyleLi, Qingxin, and Congbao Kang. 2021. "Structure and Dynamics of Zika Virus Protease and Its Insights into Inhibitor Design" Biomedicines 9, no. 8: 1044. https://doi.org/10.3390/biomedicines9081044
APA StyleLi, Q., & Kang, C. (2021). Structure and Dynamics of Zika Virus Protease and Its Insights into Inhibitor Design. Biomedicines, 9(8), 1044. https://doi.org/10.3390/biomedicines9081044





