Use of MALDI Mass Spectrometry Imaging to Identify Proteomic Signatures in Aortic Aneurysms after Endovascular Repair
Abstract
:1. Introduction
2. Experimental Section
2.1. Patient and Sample Cohort
2.2. MALDI-MSI
2.3. Protein Identification by Electrospray Ionization Tandem Mass Spectrometry
2.4. MALDI-MSI Data Processing for Statistical Analyses
2.5. Statistical Data Analysis
3. Results
3.1. Clinical Characterization
3.2. MALDI-MSI Data
3.3. Discriminative Proteins from TAA, AAA, and EVAR Tissue Sections Based on MALDI-MSI Data
4. Discussion
4.1. Summary
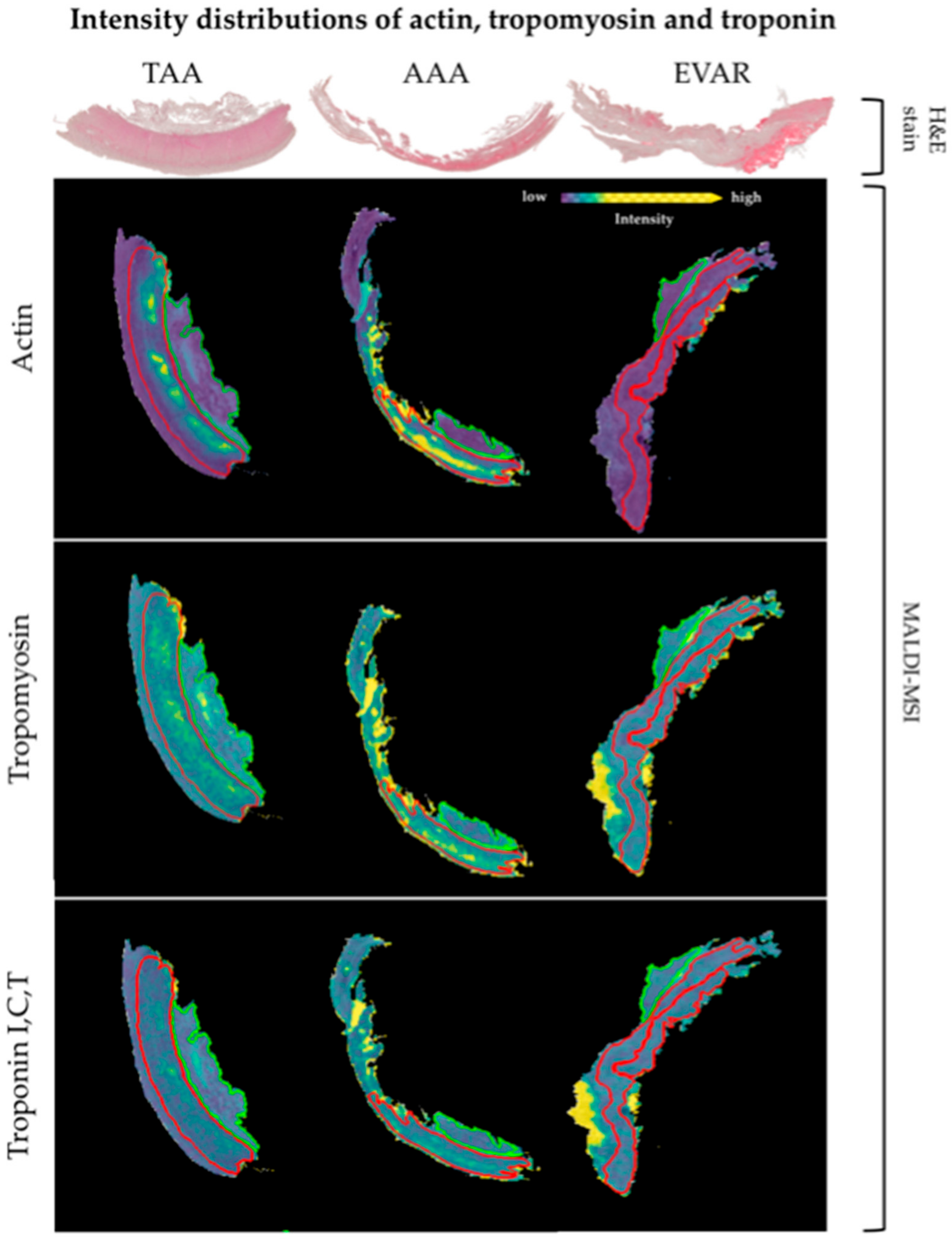
4.2. Impaired Vascular Smooth Muscle Cell Contractility after EVAR
4.3. Increased ECM Degradation after EVAR
4.4. Alteration of the Energy Supply after EVAR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nordon, I.M.; Hinchliffe, R.J.; Loftus, I.M.; Thompson, M.M. Pathophysiology and epidemiology of abdominal aortic aneurysms. Nat. Rev. Cardiol. 2011, 8, 92–102. [Google Scholar] [CrossRef]
- Ruddy, J.M.; Jones, J.A.; Spinale, F.G.; Ikonomidis, J.S. Regional heterogeneity within the aorta: Relevance to aneurysm disease. J. Thorac. Cardiovasc. Surg. 2008, 136, 1123–1130. [Google Scholar] [CrossRef] [Green Version]
- Boddy, A.M.; Lenk, G.M.; Lillvis, J.H.; Nischan, J.; Kyo, Y.; Kuivaniemi, H. Basic research studies to understand aneurysm disease. Drug News Perspect. 2008, 21, 142–148. [Google Scholar]
- Wassef, M.; Baxter, B.T.; Chisholm, R.L.; Dalman, R.L.; Fillinger, M.F.; Heinecke, J.; Humphrey, J.D.; Kuivaniemi, H.; Parks, W.C.; Pearce, W.H.; et al. Pathogenesis of abdominal aortic aneurysms: A multidisciplinary research program supported by the National Heart, Lung, and Blood Institute. J. Vasc. Surg. 2001, 34, 730–738. [Google Scholar] [CrossRef] [Green Version]
- Parodi, J.C.; Palmaz, J.C.; Barone, H.D. Transfemoral Intraluminal Graft Implantation for Abdominal Aortic Aneurysms. Ann. Vasc. Surg. 1991, 5, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Greenhalgh, R.M.; Brown, L.C.; Kwong, G.P.; Powell, J.T.; Thompson, S.G. Comparison of endovascular aneurysm repair with open repair in patients with abdominal aortic aneurysm (EVAR trial 1), 30-day operative mortality results: Randomised controlled trial. Lancet 2004, 364, 843–848. [Google Scholar] [CrossRef]
- Prinssen, M.; Verhoeven, E.L.; Buth, J.D.; Cuypers, P.W.; Van Sambeek, M.R.; Balm, R.; Buskens, E.; Grobbee, D.E.; Blankensteijn, J. A Randomized Trial Comparing Conventional and Endovascular Repair of Abdominal Aortic Aneurysms. N. Engl. J. Med. 2004, 351, 1607–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lederle, F.A.; Freischlag, J.A.; Kyriakides, T.C.; Padberg, F.T., Jr.; Matsumura, J.S.; Kohler, T.R.; Lin, P.H.; Jean-Claude, J.M.; Cikrit, D.F.; Swanson, K.M.; et al. Outcomes Following Endovascular vs Open Repair of Abdominal Aortic AneurysmA Randomized Trial. JAMA 2009, 302, 1535–1542. [Google Scholar] [CrossRef] [Green Version]
- Greenhalgh, R.M.; Brown, L.C.; Powussell, J.T.; Thompson, S.G.; Epstein, D.; Sculpher, M.J. Endovascular versus Open Repair of Abdominal Aortic Aneurysm. N. Engl. J. Med. 2010, 362, 1863–1871. [Google Scholar] [CrossRef] [Green Version]
- Lederle, F.A.; Kyriakides, T.C.; Stroupe, K.T.; Freischlag, J.A.; Padberg, F.T., Jr.; Matsumura, J.S.; Huo, Z.; Johnson, G.R. Open versus Endovascular Repair of Abdominal Aortic Aneurysm. N. Engl. J. Med. 2019, 380, 2126–2135. [Google Scholar] [CrossRef] [PubMed]
- Lal, B.K.; Zhou, W.; Li, Z.; Kyriakides, T.; Matsumura, J.; Lederle, F.A.; Freischlag, J. Predictors and outcomes of endoleaks in the Veterans Affairs Open Versus Endovascular Repair (OVER) Trial of Abdominal Aortic Aneurysms. J. Vasc. Surg. 2015, 62, 1394–1404. [Google Scholar] [CrossRef] [Green Version]
- Mehta, M.; Sternbach, Y.; Taggert, J.B.; Kreienberg, P.B.; Roddy, S.P.; Paty, P.S.; Ozsvath, K.J.; Darling, R.C., 3rd. Long-term outcomes of secondary procedures after endovascular aneurysm repair. J. Vasc. Surg. 2010, 52, 1442–1449. [Google Scholar] [CrossRef] [Green Version]
- Patel, R.; Sweeting, M.J.; Powell, J.T.; Greenhalgh, R.M. Endovascular versus open repair of abdominal aortic aneurysm in 15-years’ follow-up of the UK endovascular aneurysm repair trial 1 (EVAR trial 1): A randomised controlled trial. Lancet 2016, 388, 2366–2374. [Google Scholar] [CrossRef] [Green Version]
- Menges, A.-L.; Busch, A.; Reutersberg, B.; Trenner, M.; Kath, P.; Chernogubova, E.; Maegdefessel, L.; Eckstein, H.-H.; Zimmermann, A. The structural atrophy of the aneurysm wall in secondary expanding aortic aneurysms with endoleak type II. J. Vasc. Surg. 2019, 70, 1318–1326.e1315. [Google Scholar] [CrossRef]
- Petsophonsakul, P.; Furmanik, M.; Forsythe, R.; Dweck, M.; Schurink, G.W.; Natour, E.; Reutelingsperger, C.; Jacobs, M.; Mees, B.; Schurgers, L. Role of Vascular Smooth Muscle Cell Phenotypic Switching and Calcification in Aortic Aneurysm Formation. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1351–1368. [Google Scholar] [CrossRef]
- Jana, S.; Hu, M.; Shen, M.; Kassiri, Z. Extracellular matrix, regional heterogeneity of the aorta, and aortic aneurysm. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Milewicz, D.M.; Trybus, K.M.; Guo, D.-C.; Sweeney, H.L.; Regalado, E.; Kamm, K.; Stull, J.T. Altered Smooth Muscle Cell Force Generation as a Driver of Thoracic Aortic Aneurysms and Dissections. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 26–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milewicz, D.M.; Guo, D.-C.; Tran-Fadulu, V.; Lafont, A.L.; Papke, C.L.; Inamoto, S.; Kwartler, C.S.; Pannu, H. Genetic Basis of Thoracic Aortic Aneurysms and Dissections: Focus on Smooth Muscle Cell Contractile Dysfunction. Annu. Rev. Genom. Hum. Genet. 2008, 9, 283–302. [Google Scholar] [CrossRef] [PubMed]
- Bogunovic, N.; Meekel, J.P.; Micha, D.; Blankensteijn, J.D.; Hordijk, P.L.; Yeung, K.K. Impaired smooth muscle cell contractility as a novel concept of abdominal aortic aneurysm pathophysiology. Sci. Rep. 2019, 9, 6837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohlendieck, K. Skeletal muscle proteomics: Current approaches, technical challenges and emerging techniques. Skelet. Muscle 2011, 1, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, C.; Carberry, S.; Ohlendieck, K. Proteomic profiling of x-linked muscular dystrophy. J. Muscle Res. Cell Motil. 2009, 30, 267–269. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. Quantitative, High-Resolution Proteomics for Data-Driven Systems Biology. Annu. Rev. Biochem. 2011, 80, 273–299. [Google Scholar] [CrossRef]
- Anderson, N.L.; Anderson, N.G. The Human Plasma Proteome: History, character, and diagnostic prospects. Mol. Cell. Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maerkens, A.; Kley, R.A.; Olivé, M.; Theis, V.; van der Ven, P.F.; Reimann, J.; Milting, H.; Schreiner, A.; Uszkoreit, J.; Eisenacher, M.; et al. Differential proteomic analysis of abnormal intramyoplasmic aggregates in desminopathy. J. Proteom. 2013, 90, 14–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aichler, M.; Walch, A. MALDI Imaging mass spectrometry: Current frontiers and perspectives in pathology research and practice. Lab. Investig. 2015, 95, 422–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, S.A.; Taube, E.T.; Thiele, H.; Noack, F.; Nebrich, G.; Mohamady, K.; Hanke, T.; Klein, O. Evaluation of the Aortopathy in the Ascending Aorta: The Novelty of Using Matrix-Assisted Laser Desorption/Ionization Imaging. Proteom. Clin. Appl. 2020, 15, e2000047. [Google Scholar] [CrossRef] [PubMed]
- Dilillo, M.; Ait-Belkacem, R.; Esteve, C.; Pellegrini, D.; Nicolardi, S.; Costa, M.; Vannini, E.; Graaf, E.L.; Caleo, M.; McDonnell, L.A. Ultra-High Mass Resolution MALDI Imaging Mass Spectrometry of Proteins and Metabolites in a Mouse Model of Glioblastoma. Sci. Rep. 2017, 7, 603. [Google Scholar] [CrossRef] [Green Version]
- Mezger, S.T.P.; Mingels, A.M.A.; Bekers, O.; Heeren, R.M.A.; Cillero-Pastor, B. Mass Spectrometry Spatial-Omics on a Single Conductive Slide. Anal. Chem. 2021, 93, 2527–2533. [Google Scholar] [CrossRef]
- Spraggins, J.M.; Rizzo, D.G.; Moore, J.L.; Noto, M.J.; Skaar, E.P.; Caprioli, R.M. Next-generation technologies for spatial proteomics: Integrating ultra-high speed MALDI-TOF and high mass resolution MALDI FTICR imaging mass spectrometry for protein analysis. Proteomics 2016, 16, 1678–1689. [Google Scholar] [CrossRef] [Green Version]
- Klein, O.; Kanter, F.; Kulbe, H.; Jank, P.; Denkert, C.; Nebrich, G.; Schmitt, W.D.; Wu, Z.; Kunze, C.A.; Sehouli, J.; et al. MALDI-Imaging for Classification of Epithelial Ovarian Cancer Histotypes from a Tissue Microarray Using Machine Learning Methods. Proteom. Clin. Appl. 2019, 13, e1700181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascini, N.E.; Teunissen, J.; Noorlag, R.; Willems, S.M.; Heeren, R.M. Tumor classification with MALDI-MSI data of tissue microarrays: A case study. Methods 2018, 151, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Klein, O.; Fogt, F.; Hollerbach, S.; Nebrich, G.; Boskamp, T.; Wellmann, A. Classification of Inflammatory Bowel Disease from Formalin-Fixed, Paraffin-Embedded Tissue Biopsies via Imaging Mass Spectrometry. Proteom. Clin. Appl. 2020, 14, 1900131. [Google Scholar] [CrossRef]
- Klein, O.; Strohschein, K.; Nebrich, G.; Oetjen, J.; Trede, D.; Thiele, H.; Alexandrov, T.; Giavalisco, P.; Duda, G.N.; Von Roth, P.; et al. MALDI imaging mass spectrometry: Discrimination of pathophysiological regions in traumatized skeletal muscle by characteristic peptide signatures. Proteomics 2014, 14, 2249–2260. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ly, A.; Longuespée, R.; Casadonte, R.; Wandernoth, P.; Schwamborn, K.; Bollwein, C.; Marsching, C.; Kriegsmann, K.; Hopf, C.; Weichert, W.; et al. Site-to-Site Reproducibility and Spatial Resolution in MALDI–MSI of Peptides from Formalin-Fixed Paraffin-Embedded Samples. Proteom. Clin. Appl. 2019, 13, e1800029. [Google Scholar] [CrossRef] [Green Version]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Cillero-Pastor, B.; Heeren, R.M. Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging for Peptide and Protein Analyses: A Critical Review of On-Tissue Digestion. J. Proteome Res. 2014, 13, 325–335. [Google Scholar] [CrossRef]
- Alexandrov, T.; Becker, M.; Deininger, S.O.; Ernst, G.; Wehder, L.; Grasmair, M.; von Eggeling, F.; Thiele, H.; Maass, P. Spatial Segmentation of Imaging Mass Spectrometry Data with Edge-Preserving Image Denoising and Clustering. J. Proteome Res. 2010, 9, 6535–6546. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, T.; Becker, M.; Guntinas-Lichius, O.; Ernst, G.; von Eggeling, F. MALDI-imaging segmentation is a powerful tool for spatial functional proteomic analysis of human larynx carcinoma. J. Cancer Res. Clin. Oncol. 2012, 139, 85–95. [Google Scholar] [CrossRef]
- Trede, D.; Schiffler, S.; Becker, M.; Wirtz, S.; Steinhorst, K.; Strehlow, J.; Aichler, M.; Kobarg, J.H.; Oetjen, J.; Dyatlov, A.; et al. Exploring Three-Dimensional Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry Data: Three-Dimensional Spatial Segmentation of Mouse Kidney. Anal. Chem. 2012, 84, 6079–6087. [Google Scholar] [CrossRef]
- Marston, S.; El-Mezgueldi, M. Role of tropomyosin in the regulation of contraction in smooth muscle. Adv. Exp. Med. Biol. 2008, 644, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Moran, C.M.; Garriock, R.J.; Miller, M.K.; Heimark, R.L.; Gregorio, C.C.; Krieg, P.A. Expression of the fast twitch troponin complex, fTnT, fTnI and fTnC, in vascular smooth muscle. Cell Motil. Cytoskelet. 2008, 65, 652–661. [Google Scholar] [CrossRef] [Green Version]
- Kajioka, S.; Takahashi-Yanaga, F.; Shahab, N.; Onimaru, M.; Matsuda, M.; Takahashi, R.; Asano, H.; Morita, H.; Morimoto, S.; Yonemitsu, Y.; et al. Endogenous Cardiac Troponin T Modulates (Ca2+)—Mediated Smooth Muscle Contraction. Sci. Rep. 2012, 2, 979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagenseil, J.E.; Mecham, R.P. Vascular Extracellular Matrix and Arterial Mechanics. Physiol. Rev. 2009, 89, 957–989. [Google Scholar] [CrossRef] [Green Version]
- Clyman, R.I.; McDonald, K.A.; Kramer, R.H. Integrin receptors on aortic smooth muscle cells mediate adhesion to fibronectin, laminin, and collagen. Circ. Res. 1990, 67, 175–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carmo, M.; Colombo, L.; Bruno, A.; Corsi, F.R.; Roncoroni, L.; Cuttin, M.S.; Radice, F.; Mussini, E.; Settembrini, P.G. Alteration of Elastin, Collagen and their Cross-links in Abdominal Aortic Aneurysms. Eur. J. Vasc. Endovasc. Surg. 2002, 23, 543–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blassova, T.; Tonar, Z.; Tomasek, P.; Hosek, P.; Hollan, I.; Treska, V.; Molacek, J. Inflammatory cell infiltrates, hypoxia, vascularization, pentraxin 3 and osteoprotegerin in abdominal aortic aneurysms—A quantitative histological study. PLoS ONE 2019, 14, e0224818. [Google Scholar] [CrossRef]
- Keeling, W.B.; Armstrong, P.A.; Stone, P.A.; Bandyk, D.F.; Shames, M.L. An Overview of Matrix Metalloproteinases in the Pathogenesis and Treatment of Abdominal Aortic Aneurysms. Vasc. Endovasc. Surg. 2005, 39, 457–464. [Google Scholar] [CrossRef]
- Lee, C.-C.; Lee, M.-T.G.; Chen, Y.-S.; Lee, S.-H.; Chen, Y.-S.; Chen, S.-C.; Chang, S.-C. Risk of Aortic Dissection and Aortic Aneurysm in Patients Taking Oral Fluoroquinolone. JAMA Intern. Med. 2015, 175, 1839–1847. [Google Scholar] [CrossRef]
- Ailawadi, G.; Moehle, C.W.; Pei, H.; Walton, S.P.; Yang, Z.; Kron, I.L.; Lau, C.L.; Owens, G.K. Smooth muscle phenotypic modulation is an early event in aortic aneurysms. J. Thorac. Cardiovasc. Surg. 2009, 138, 1392–1399. [Google Scholar] [CrossRef] [Green Version]
- López-Candales, A.; Holmes, D.R.; Liao, S.; Scott, M.J.; Wickline, S.A.; Thompson, R.W. Decreased vascular smooth muscle cell density in medial degeneration of human abdominal aortic aneurysms. Am. J. Pathol. 1997, 150, 993–1007. [Google Scholar]
- Jiao, L.; Xu, Z.; Xu, F.; Zhang, S.; Wu, K. Vascular smooth muscle cell remodelling in elastase-induced aortic aneurysm. Acta Cardiol. 2010, 65, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Toogood, H.S.; Leys, D.; Scrutton, N.S. Dynamics driving function − new insights from electron transferring flavoproteins and partner complexes. FEBS J. 2007, 274, 5481–5504. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.L.; Frerman, F.E.; Kim, J.J. Three-dimensional structure of human electron transfer flavoprotein to 2.1—A resolution. Proc. Natl. Acad. Sci. USA 1996, 93, 14355–14360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chohan, K.K.; Jones, M.; Grossmann, J.G.; Frerman, F.E.; Scrutton, N.S.; Sutcliffe, M.J. Protein Dynamics Enhance Electronic Coupling in Electron Transfer Complexes. J. Biol. Chem. 2001, 276, 34142–34147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frerman, F.E. Acyl-CoA dehydrogenases, electron transfer flavoprotein and electron transfer flavoprotein dehydrogenase. Biochem. Soc. Trans. 1988, 16, 416–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckmann, J.D.; Frerman, F.E. Electron transfer flavoprotein-ubiquinone oxidoreductase from pig liver: Purification and molecular, redox, and catalytic properties. Biochemistry 1985, 24, 3913–3921. [Google Scholar] [CrossRef]
- Wolinsky, H.; Glagov, S. Comparison of Abdominal and Thoracic Aortic Medial Structure in Mammals. Deviation of man from the usual pattern. Circ. Res. 1969, 25, 677–686. [Google Scholar] [CrossRef] [Green Version]

| Subgroup | Gender | Age | Maximum Aneurysm Diameter (mm) | Comorbidities |
|---|---|---|---|---|
| TAA_1 | M | 62 | 58 | CAD, HI |
| TAA_2 | M | 67 | 59 | HI |
| TAA_3 | M | 54 | 52 | HI |
| TAA_4 | M | 43 | 57 | HI |
| TAA_5 | M | 68 | 46 | CAD, HI, AHT, HLP, COPD, CRF |
| AAA_1 | M | 78 | 60 | CAD, AHT, HLP |
| AAA_2 | M | 72 | 81 | CRF, AHT |
| AAA_3 | M | 78 | 80 | CAD, HLP |
| AAA_4 | M | 63 | 110 | AHT |
| AAA_5 | M | 76 | 68 | CAD, HI, AHT |
| EVAR_1 | M | 67 | 75 | CAD, PAD, AHT |
| EVAR_2 | M | 64 | 90 | CAD, AHT, HLP |
| EVAR_3 | M | 80 | 65 | AF, PAD |
| EVAR_4 | M | 73 | 59 | CAD, CRF |
| EVAR_5 | M | 65 | 69 | AHT, PAD |
| MALDI-MSI m/z Value [Da] | ROC [AUC] Media AAA vs. EVAR | ROC [AUC] Media AAA vs. TAA | ROC [AUC] Media EVAR vs. TAA | LC-MS [MH + Calc.] | Scores | Sequence | Gene Symbol | Protein |
|---|---|---|---|---|---|---|---|---|
| 976.43 | 0.62 | 0.35 | 0.22 | 976.4468194 | 88.56 | AGFAGDDAPR | ACTA2 | Actin, aortic smooth muscle |
| 1198.65 | 0.66 | 0.29 | 0.10 | 1198.51941 | 57.19 | DSYVGDEAQSK | ||
| 1198.65 | 0.66 | 0.29 | 0.10 | 1198.703002 | 41.77 | AVFPSIVGRPR | ||
| 1580.684 | 0.56 | 0.45 | 0.39 | 1580.800411 | 43.04 | MQKEITALAPSTMK | ||
| 1790.904 | 0.57 | 0.36 | 0.27 | 1790.891339 | 28.06 | SYELPDGQVITIGNER | ||
| 1564.883 | 0.59 | 0.42 | 0.33 | 1564.905355 | 31.49 | SIVVSPILIPENQR | CDH13 | Cadherin-13 |
| 1835.908 | 0.58 | 0.44 | 0.34 | 1836.842546 | 32.84 | MTAFDADDPATDNALLR | ||
| 836.417 | 0.59 | 0.42 | 0.31 | 836.4361428 | 49.08 | GPAGPQGPR | COL1A1 | Collagen alpha-1(I) chain |
| 852.418 | 0.61 | 0.40 | 0.28 | 851.4249814 | 43.88 | GFSGLDGAK | ||
| 868.42 | 0.59 | 0.42 | 0.31 | 868.4253882 | 34.63 | GEAGPQGPR | ||
| 886.421 | 0.56 | 0.43 | 0.36 | 886.4359955 | 47.39 | GSEGPQGVR | ||
| 784.412 | 0.58 | 0.45 | 0.37 | 785.3889089 | 48.12 | GDQGPVGR | COL1A2 | Collagen alpha-2(I) chain |
| 1561.883 | 0.60 | 0.48 | 0.37 | 1562.79067 | 87.57 | GETGPSGPVGPAGAVGPR | ||
| 1237.653 | 0.57 | 0.43 | 0.37 | 1238.651334 | 47.18 | VAVFFSNTPTR | COL6A3 | Collagen alpha-3(VI) chain |
| 1459.673 | 0.61 | 0.43 | 0.32 | 1459.860756 | 25.39 | IGDLHPQIVNLLK | ||
| 1462.674 | 0.55 | 0.44 | 0.39 | 1462.763096 | 27.26 | QINVGNALEYVSR | ||
| 1508.678 | 0.60 | 0.40 | 0.32 | 1507.799786 | 83.26 | LSVEALNSLTGEFK | CKM | Creatine kinase M-type Desmin |
| 1508.678 | 0.60 | 0.40 | 0.32 | 1507.699725 | 58.36 | GGDDLDPNYVLSSR | ||
| 1767.902 | 0.57 | 0.47 | 0.39 | 1768.83488 | 32.75 | DGEVVSEATQQQHEVL | ||
| 2088.931 | 0.56 | 0.45 | 0.39 | 2088.091512 | 51.57 | TFGGAPGFPLGSPLSSPVFPR | ||
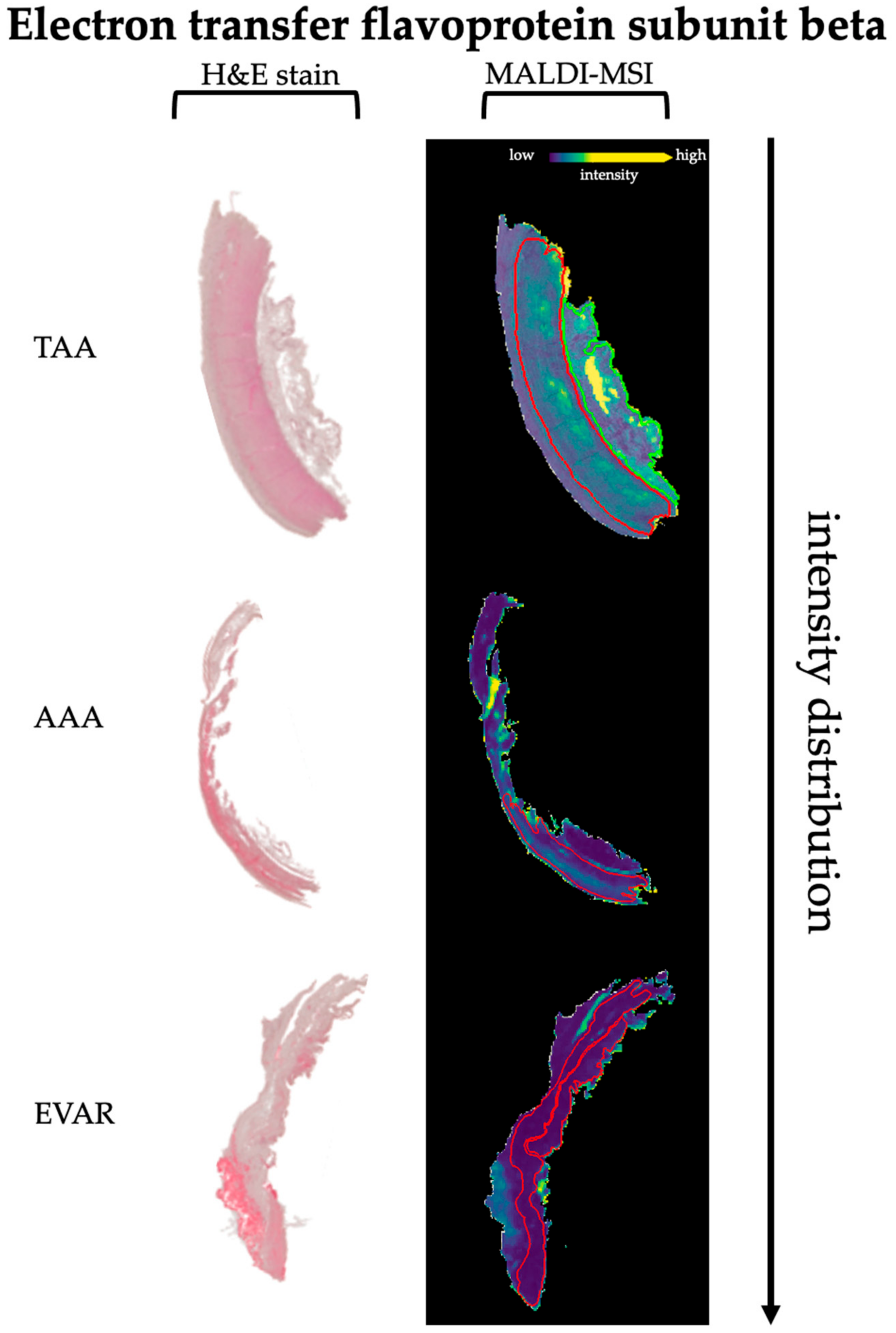
| 853.418 | 0.61 | 0.40 | 0.27 | 853.5233919 | 55.31 | LGPLQVAR | ETFB | Electron transfer flavoprotein subunit beta |
| 1340.663 | 0.60 | 0.44 | 0.34 | 1339.720602 | 51.07 | LSVISVEDPPQR | ||
| 974.429 | 0.56 | 0.43 | 0.38 | 973.6021152 | 45.36 | SGVSLAALKK | H1-3 | Histone H1.3 |
| 1106.641 | 0.58 | 0.42 | 0.32 | 1107.565851 | 47.87 | ALAAAGYDVEK | ||
| 1198.65 | 0.66 | 0.29 | 0.10 | 1198.666651 | 64.14 | ASGPPVSELITK | ||
| 1325.661 | 0.59 | 0.44 | 0.36 | 1325.752447 | 45.61 | DNIQGITKPAIR | H4C1 | Histone H4 |
| 1466.674 | 0.60 | 0.42 | 0.32 | 1466.801839 | 61.03 | TVTAMDVVYALKR | ||
| 1533.68 | 0.58 | 0.44 | 0.34 | 1533.775171 | 82.04 | VVDSLQTSLDAETR | MYO6 | Myosin-6 |
| 1850.909 | 0.59 | 0.44 | 0.34 | 1851.041427 | 48.84 | VQLLHSQNTSLINQKK | ||
| 2088.931 | 0.56 | 0.45 | 0.39 | 2088.123001 | 33.09 | YRILNPVAIPEGQFIDSR | ||
| 2199.941 | 0.57 | 0.46 | 0.39 | 2200.123705 | 49.68 | GTLEDQIIQANPALEAFGNAK | ||
| 976.43 | 0.62 | 0.35 | 0.22 | 975.4887038 | 36.63 | AMEAVAAQGK | PGAM2 | Phosphoglycerate mutase 2 |
| 1150.645 | 0.56 | 0.43 | 0.38 | 1150.666958 | 45.55 | VLIAAHGNSLR | ||
| 875.42 | 0.57 | 0.44 | 0.37 | 875.4465169 | 30.27 | SLEAQAEK | TPM1 | Tropomyosin alpha-1 chain |
| 1460.674 | 0.62 | 0.43 | 0.31 | 1460.731208 | 39.4 | KATDAEADVASLNR | ||
| 1516.679 | 0.61 | 0.43 | 0.32 | 1516.819568 | 27.34 | SKQLEDELVSLQK | ||
| 1305.659 | 0.58 | 0.41 | 0.34 | 1306.638768 | 44.38 | KNIDALSGMEGR | TNNI3 | Troponin I, cardiac muscle |
| 1479.675 | 0.54 | 0.43 | 0.40 | 1479.727686 | 42.27 | ISADAMMQALLGAR | ||
| 1889.913 | 0.57 | 0.46 | 0.38 | 1890.031221 | 44.46 | NITEIADLTQKIFDLR | ||
| 758.41 | 0.55 | 0.45 | 0.40 | 757.4673223 | 35.39 | ILAERR | TNNT2 | Troponin T, cardiac muscle |
| 906.423 | 0.55 | 0.43 | 0.37 | 906.5021046 | 26.65 | YEINVLR | ||
| 1797.904 | 0.57 | 0.46 | 0.40 | 1796.934971 | 29.11 | SFMPNLVPPKIPDGER | ||
| 1143.445 | 0.60 | 0.42 | 0.31 | 1143.632775 | 28.94 | LAVNMVPFPR | TUBB | Tubulin beta chain |
| 1320.661 | 0.64 | 0.38 | 0.24 | 1319.701066 | 49.82 | IMNTFSVVPSPK | ||
| 1269.656 | 0.56 | 0.43 | 0.37 | 1270.559399 | 37.39 | LGDLYEEEMR | VIM | Vimentin |
| 1428.671 | 0.55 | 0.33 | 0.28 | 1428.710851 | 40.41 | SLYASSPGGVYATR | ||
| 2498.168 | 0.55 | 0.49 | 0.44 | 2497.256473 | 42.86 | LLQDSVDFSLADAINTEFKNTR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buerger, M.; Klein, O.; Kapahnke, S.; Mueller, V.; Frese, J.P.; Omran, S.; Greiner, A.; Sommerfeld, M.; Kaschina, E.; Jannasch, A.; et al. Use of MALDI Mass Spectrometry Imaging to Identify Proteomic Signatures in Aortic Aneurysms after Endovascular Repair. Biomedicines 2021, 9, 1088. https://doi.org/10.3390/biomedicines9091088
Buerger M, Klein O, Kapahnke S, Mueller V, Frese JP, Omran S, Greiner A, Sommerfeld M, Kaschina E, Jannasch A, et al. Use of MALDI Mass Spectrometry Imaging to Identify Proteomic Signatures in Aortic Aneurysms after Endovascular Repair. Biomedicines. 2021; 9(9):1088. https://doi.org/10.3390/biomedicines9091088
Chicago/Turabian StyleBuerger, Matthias, Oliver Klein, Sebastian Kapahnke, Verena Mueller, Jan Paul Frese, Safwan Omran, Andreas Greiner, Manuela Sommerfeld, Elena Kaschina, Anett Jannasch, and et al. 2021. "Use of MALDI Mass Spectrometry Imaging to Identify Proteomic Signatures in Aortic Aneurysms after Endovascular Repair" Biomedicines 9, no. 9: 1088. https://doi.org/10.3390/biomedicines9091088
APA StyleBuerger, M., Klein, O., Kapahnke, S., Mueller, V., Frese, J. P., Omran, S., Greiner, A., Sommerfeld, M., Kaschina, E., Jannasch, A., Dittfeld, C., Mahlmann, A., & Hinterseher, I. (2021). Use of MALDI Mass Spectrometry Imaging to Identify Proteomic Signatures in Aortic Aneurysms after Endovascular Repair. Biomedicines, 9(9), 1088. https://doi.org/10.3390/biomedicines9091088







