Epigenetic Mechanisms Underlying COVID-19 Pathogenesis
Abstract
:1. Introduction
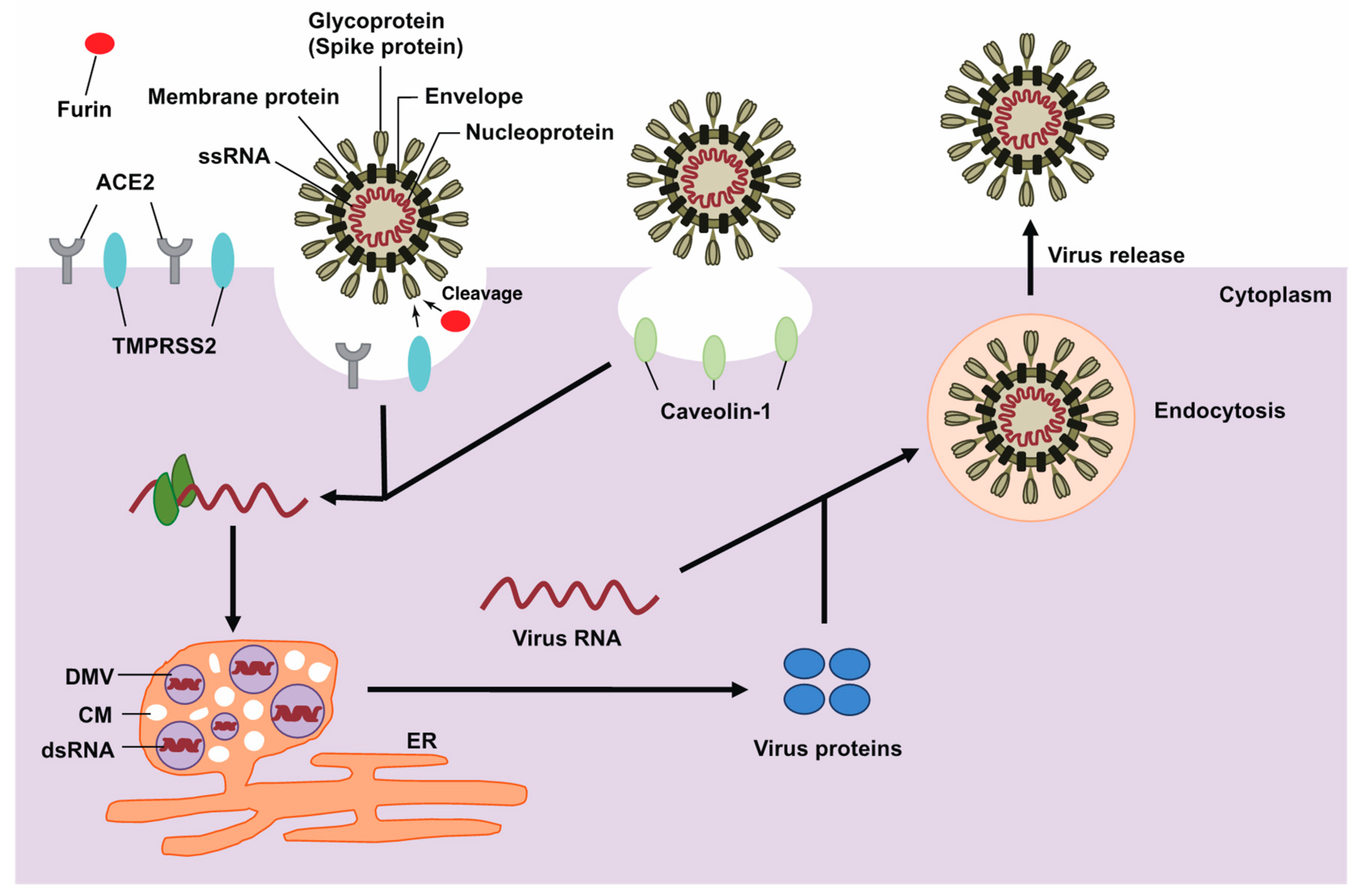
2. The Life Cycle of SARS-CoV-2
3. RNA Modifications of the SARS-CoV-2 Genome
4. ACE2 Gene Expression
5. Effect of DNA Methylation on ACE2 Expression
6. Alteration of DNA Methylation in Patients with COVID-19
7. Histone Modifications Related to ACE2 Gene Expression and COVID-19
8. ACE2 Gene Regulation Mediated by miRNAs, lncRNAs, and circRNAs
9. Identification of SARS-CoV-2 Infection Factors Using Genome-Wide Screening Analysis
10. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Islam, M.A.; Kundu, S.; Alam, S.S.; Hossan, T.; Kamal, M.A.; Hassan, R. Prevalence and characteristics of fever in adult and paediatric patients with coronavirus disease 2019 (COVID-19): A systematic review and meta-analysis of 17,515 patients. PLoS ONE 2021, 16, e0249788. [Google Scholar] [CrossRef]
- Islam, M.A.; Alam, S.S.; Kundu, S.; Hossan, T.; Kamal, M.A.; Cavestro, C. Prevalence of Headache in Patients with Coronavirus Disease 2019 (COVID-19): A Systematic Review and Meta-Analysis of 14,275 Patients. Front. Neurol. 2020, 11, 562634. [Google Scholar] [CrossRef]
- Saniasiaya, J.; Islam, M.A.; Abdullah, B. Prevalence of Olfactory Dysfunction in Coronavirus Disease 2019 (COVID-19): A Meta-analysis of 27,492 Patients. Laryngoscope 2021, 131, 865–878. [Google Scholar] [CrossRef]
- Saniasiaya, J.; Islam, M.A.; Abdullah, B. Prevalence and Characteristics of Taste Disorders in Cases of COVID-19: A Meta-analysis of 29,349 Patients. Otolaryngol. Head Neck Surg. 2021, 165, 33–42. [Google Scholar] [CrossRef]
- Agyeman, A.A.; Chin, K.L.; Landersdorfer, C.B.; Liew, D.; Ofori-Asenso, R. Smell and Taste Dysfunction in Patients With COVID-19: A Systematic Review and Meta-analysis. Mayo Clin. Proc. 2020, 95, 1621–1631. [Google Scholar] [CrossRef]
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef]
- Williams, G.H.; Llewelyn, A.; Brandao, R.; Chowdhary, K.; Hardisty, K.M.; Loddo, M. SARS-CoV-2 testing and sequencing for international arrivals reveals significant cross border transmission of high risk variants into the United Kingdom. EClinicalMedicine 2021, 38, 101021. [Google Scholar] [CrossRef]
- Cascella, M.; Rajnik, M.; Aleem, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Le, T.T.; Cramer, J.P.; Chen, R.; Mayhew, S. Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020, 19, 667–668. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Founou, R.C.; Blocker, A.J.; Noubom, M.; Tsayem, C.; Choukem, S.P.; Dongen, M.V.; Founou, L.L. The COVID-19 pandemic: A threat to antimicrobial resistance containment. Future Sci. OA 2021, 7, FSO736. [Google Scholar] [CrossRef] [PubMed]
- Siemieniuk, R.A.; Bartoszko, J.J.; Ge, L.; Zeraatkar, D.; Izcovich, A.; Kum, E.; Pardo-Hernandez, H.; Qasim, A.; Martinez, J.P.D.; Rochwerg, B.; et al. Drug treatments for COVID-19: Living systematic review and network meta-analysis. BMJ 2020, 370, m2980. [Google Scholar] [CrossRef]
- Chatterjee, S.; Mishra, S.; Chowdhury, K.D.; Ghosh, C.K.; Saha, K.D. Various theranostics and immunization strategies based on nanotechnology against COVID-19 pandemic: An interdisciplinary view. Life Sci. 2021, 278, 119580. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, R.; Komatsu, M.; Takasawa, K.; Asada, K.; Kaneko, S. Epigenetics Analysis and Integrated Analysis of Multiomics Data, Including Epigenetic Data, Using Artificial Intelligence in the Era of Precision Medicine. Biomolecules 2020, 10, 62. [Google Scholar] [CrossRef] [Green Version]
- Kogure, M.; Takawa, M.; Saloura, V.; Sone, K.; Piao, L.; Ueda, K.; Ibrahim, R.; Tsunoda, T.; Sugiyama, M.; Atomi, Y.; et al. The oncogenic polycomb histone methyltransferase EZH2 methylates lysine 120 on histone H2B and competes ubiquitination. Neoplasia 2013, 15, 1251–1261. [Google Scholar] [CrossRef] [Green Version]
- Nakakido, M.; Deng, Z.; Suzuki, T.; Dohmae, N.; Nakamura, Y.; Hamamoto, R. PRMT6 increases cytoplasmic localization of p21CDKN1A in cancer cells through arginine methylation and makes more resistant to cytotoxic agents. Oncotarget 2015, 6, 30957–30967. [Google Scholar] [CrossRef] [Green Version]
- Oki, S.; Sone, K.; Oda, K.; Hamamoto, R.; Ikemura, M.; Maeda, D.; Takeuchi, M.; Tanikawa, M.; Mori-Uchino, M.; Nagasaka, K.; et al. Oncogenic histone methyltransferase EZH2: A novel prognostic marker with therapeutic potential in endometrial cancer. Oncotarget 2017, 8, 40402–40411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.K.; Kim, K.; Ryu, J.W.; Ryu, T.Y.; Lim, J.H.; Oh, J.H.; Min, J.K.; Jung, C.R.; Hamamoto, R.; Son, M.Y.; et al. The novel prognostic marker, EHMT2, is involved in cell proliferation via HSPD1 regulation in breast cancer. Int. J. Oncol. 2019, 54, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Bolatkan, A.; Kaneko, S.; Ikawa, N.; Asada, K.; Komatsu, M.; Hayami, S.; Ojima, H.; Abe, N.; Yamaue, H.; et al. Deregulation of the Histone Lysine-Specific Demethylase 1 Is Involved in Human Hepatocellular Carcinoma. Biomolecules 2019, 9, 810. [Google Scholar] [CrossRef] [Green Version]
- Ryu, J.W.; Kim, S.K.; Son, M.Y.; Jeon, S.J.; Oh, J.H.; Lim, J.H.; Cho, S.; Jung, C.R.; Hamamoto, R.; Kim, D.S.; et al. Novel prognostic marker PRMT1 regulates cell growth via downregulation of CDKN1A in HCC. Oncotarget 2017, 8, 115444–115455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saloura, V.; Cho, H.S.; Kyiotani, K.; Alachkar, H.; Zuo, Z.; Nakakido, M.; Tsunoda, T.; Seiwert, T.; Lingen, M.; Licht, J.; et al. WHSC1 Promotes Oncogenesis through Regulation of NIMA-related-kinase-7 in Squamous Cell Carcinoma of the Head and Neck. Mol. Cancer Res. 2015, 13, 293–304. [Google Scholar] [CrossRef] [Green Version]
- Hayami, S.; Kelly, J.D.; Cho, H.S.; Yoshimatsu, M.; Unoki, M.; Tsunoda, T.; Field, H.I.; Neal, D.E.; Yamaue, H.; Ponder, B.A.; et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int. J. Cancer 2011, 128, 574–586. [Google Scholar] [CrossRef]
- Yoshimatsu, M.; Toyokawa, G.; Hayami, S.; Unoki, M.; Tsunoda, T.; Field, H.I.; Kelly, J.D.; Neal, D.E.; Maehara, Y.; Ponder, B.A.; et al. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int. J. Cancer 2011, 128, 562–573. [Google Scholar] [CrossRef]
- Sone, K.; Piao, L.; Nakakido, M.; Ueda, K.; Jenuwein, T.; Nakamura, Y.; Hamamoto, R. Critical role of lysine 134 methylation on histone H2AX for gamma-H2AX production and DNA repair. Nat. Commun. 2014, 5, 5691. [Google Scholar] [CrossRef] [Green Version]
- Licht, J.D.; Bennett, R.L. Leveraging epigenetics to enhance the efficacy of immunotherapy. Clin. Epigenet. 2021, 13, 115. [Google Scholar] [CrossRef]
- Asada, K.; Kaneko, S.; Takasawa, K.; Machino, H.; Takahashi, S.; Shinkai, N.; Shimoyama, R.; Komatsu, M.; Hamamoto, R. Integrated Analysis of Whole Genome and Epigenome Data Using Machine Learning Technology: Toward the Establishment of Precision Oncology. Front. Oncol. 2021, 11, 666937. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, R.; Suvarna, K.; Yamada, M.; Kobayashi, K.; Shinkai, N.; Miyake, M.; Takahashi, M.; Jinnai, S.; Shimoyama, R.; Sakai, A.; et al. Application of Artificial Intelligence Technology in Oncology: Towards the Establishment of Precision Medicine. Cancers 2020, 12, 3532. [Google Scholar] [CrossRef]
- Kaneko, S.; Mitsuyama, T.; Shiraishi, K.; Ikawa, N.; Shozu, K.; Dozen, A.; Machino, H.; Asada, K.; Komatsu, M.; Kukita, A.; et al. Genome-Wide Chromatin Analysis of FFPE Tissues Using a Dual-Arm Robot with Clinical Potential. Cancers 2021, 13, 2126. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e910. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Goh, G.K.; Dunker, A.K.; Foster, J.A.; Uversky, V.N. Shell Disorder Analysis Suggests That Pangolins Offered a Window for a Silent Spread of an Attenuated SARS-CoV-2 Precursor among Humans. J. Proteome Res. 2020, 19, 4543–4552. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pohlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e775. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirchdoerfer, R.N.; Cottrell, C.A.; Wang, N.; Pallesen, J.; Yassine, H.M.; Turner, H.L.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Pre-fusion structure of a human coronavirus spike protein. Nature 2016, 531, 118–121. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Tang, J.; Ma, Y.; Liang, X.; Yang, Y.; Peng, G.; Qi, Q.; Jiang, S.; Li, J.; Du, L.; et al. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015, 89, 6121–6125. [Google Scholar] [CrossRef] [Green Version]
- Hassan, S.S.; Ghosh, S.; Attrish, D.; Choudhury, P.P.; Aljabali, A.A.A.; Uhal, B.D.; Lundstrom, K.; Rezaei, N.; Uversky, V.N.; Seyran, M.; et al. Possible Transmission Flow of SARS-CoV-2 Based on ACE2 Features. Molecules 2020, 25, 5906. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nagata, N.; Shirato, K.; Kawase, M.; Takeda, M.; Taguchi, F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010, 84, 12658–12664. [Google Scholar] [CrossRef] [Green Version]
- Seyran, M.; Takayama, K.; Uversky, V.N.; Lundstrom, K.; Palu, G.; Sherchan, S.P.; Attrish, D.; Rezaei, N.; Aljabali, A.A.A.; Ghosh, S.; et al. The structural basis of accelerated host cell entry by SARS-CoV-2dagger. FEBS J. 2020. [Google Scholar] [CrossRef]
- Goh, G.; Foster, J.; Dunker, A.K.; Uversky, V. Computational, Experimental, and Clinical Evidence for a Specific but Peculiar Evolutionary Nature of (COVID-19) SARS-CoV-2. Preprints 2021, 2021050025. [Google Scholar]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells following viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef]
- Inoue, Y.; Tanaka, N.; Tanaka, Y.; Inoue, S.; Morita, K.; Zhuang, M.; Hattori, T.; Sugamura, K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007, 81, 8722–8729. [Google Scholar] [CrossRef] [Green Version]
- DeTulleo, L.; Kirchhausen, T. The clathrin endocytic pathway in viral infection. EMBO J. 1998, 17, 4585–4593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Liu, D.X.; Tam, J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008, 369, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, P.; Liu, K.; Guo, F.; Zhang, Y.; Zhang, G.; Jiang, C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008, 18, 290–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wubben, J.M.; Atkinson, S.C.; Borg, N.A. The Role of Protein Disorder in Nuclear Transport and in Its Subversion by Viruses. Cells 2020, 9, 654. [Google Scholar] [CrossRef]
- Wolff, G.; Limpens, R.; Zevenhoven-Dobbe, J.C.; Laugks, U.; Zheng, S.; de Jong, A.W.M.; Koning, R.I.; Agard, D.A.; Grunewald, K.; Koster, A.J.; et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 2020, 369, 1395–1398. [Google Scholar] [CrossRef]
- Knoops, K.; Kikkert, M.; Worm, S.H.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008, 6, e226. [Google Scholar] [CrossRef] [PubMed]
- Egger, D.; Teterina, N.; Ehrenfeld, E.; Bienz, K. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 2000, 74, 6570–6580. [Google Scholar] [CrossRef] [Green Version]
- Schlegel, A.; Giddings, T.H., Jr.; Ladinsky, M.S.; Kirkegaard, K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 1996, 70, 6576–6588. [Google Scholar] [CrossRef] [Green Version]
- Klaus, J.P.; Eisenhauer, P.; Russo, J.; Mason, A.B.; Do, D.; King, B.; Taatjes, D.; Cornillez-Ty, C.; Boyson, J.E.; Thali, M.; et al. The intracellular cargo receptor ERGIC-53 is required for the production of infectious arenavirus, coronavirus, and filovirus particles. Cell Host Microbe 2013, 14, 522–534. [Google Scholar] [CrossRef] [Green Version]
- Beske, O.; Reichelt, M.; Taylor, M.P.; Kirkegaard, K.; Andino, R. Poliovirus infection blocks ERGIC-to-Golgi trafficking and induces microtubule-dependent disruption of the Golgi complex. J. Cell Sci. 2007, 120, 3207–3218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e819. [Google Scholar] [CrossRef] [PubMed]
- Asada, K.; Bolatkan, A.; Takasawa, K.; Komatsu, M.; Kaneko, S.; Hamamoto, R. Critical Roles of N(6)-Methyladenosine (m(6)A) in Cancer and Virus Infection. Biomolecules 2020, 10, 1071. [Google Scholar] [CrossRef] [PubMed]
- Di Giorgio, S.; Martignano, F.; Torcia, M.G.; Mattiuz, G.; Conticello, S.G. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci. Adv. 2020, 6, eabb5813. [Google Scholar] [CrossRef]
- Schmidt, N.; Lareau, C.A.; Keshishian, H.; Ganskih, S.; Schneider, C.; Hennig, T.; Melanson, R.; Werner, S.; Wei, Y.; Zimmer, M.; et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat. Microbiol. 2021, 6, 339–353. [Google Scholar] [CrossRef]
- Kosuge, M.; Furusawa-Nishii, E.; Ito, K.; Saito, Y.; Ogasawara, K. Point mutation bias in SARS-CoV-2 variants results in increased ability to stimulate inflammatory responses. Sci. Rep. 2020, 10, 17766. [Google Scholar] [CrossRef]
- Wang, R.; Hozumi, Y.; Zheng, Y.H.; Yin, C.; Wei, G.W. Host Immune Response Driving SARS-CoV-2 Evolution. Viruses 2020, 12, 1095. [Google Scholar] [CrossRef]
- Donoghue, M.; Hsieh, F.; Baronas, E.; Godbout, K.; Gosselin, M.; Stagliano, N.; Donovan, M.; Woolf, B.; Robison, K.; Jeyaseelan, R.; et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000, 87, E1–E9. [Google Scholar] [CrossRef]
- Ziegler, C.G.K.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035.e19. [Google Scholar] [CrossRef]
- Bunyavanich, S.; Do, A.; Vicencio, A. Nasal Gene Expression of Angiotensin-Converting Enzyme 2 in Children and Adults. JAMA 2020, 323, 2427–2429. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Mo, X.; Hu, Y.; Qi, X.; Jiang, F.; Jiang, Z.; Tong, S. Epidemiology of COVID-19 Among Children in China. Pediatrics 2020, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senkel, S.; Lucas, B.; Klein-Hitpass, L.; Ryffel, G.U. Identification of target genes of the transcription factor HNF1beta and HNF1alpha in a human embryonic kidney cell line. Biochim. Biophys. Acta 2005, 1731, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.B.; Chhabra, K.H.; Nguyen, V.K.; Xia, H.; Lazartigues, E. The transcription factor HNF1alpha induces expression of angiotensin-converting enzyme 2 (ACE2) in pancreatic islets from evolutionarily conserved promoter motifs. Biochim. Biophys. Acta 2013, 1829, 1225–1235. [Google Scholar] [CrossRef] [Green Version]
- Onabajo, O.O.; Banday, A.R.; Stanifer, M.L.; Yan, W.; Obajemu, A.; Santer, D.M.; Florez-Vargas, O.; Piontkivska, H.; Vargas, J.M.; Ring, T.J.; et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 2020, 52, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.W.; Attig, J.; Bolland, W.; Young, G.R.; Major, J.; Wrobel, A.G.; Gamblin, S.; Wack, A.; Kassiotis, G. Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option. Nat. Genet. 2020, 52, 1294–1302. [Google Scholar] [CrossRef]
- Blume, C.; Jackson, C.L.; Spalluto, C.M.; Legebeke, J.; Nazlamova, L.; Conforti, F.; Perotin, J.M.; Frank, M.; Butler, J.; Crispin, M.; et al. A novel ACE2 isoform is expressed in human respiratory epithelia and is upregulated in response to interferons and RNA respiratory virus infection. Nat. Genet. 2021, 53, 205–214. [Google Scholar] [CrossRef]
- Salvati, L.; Biagioni, B.; Vivarelli, E.; Parronchi, P. A gendered magnifying glass on COVID-19. Clin. Mol. Allergy 2020, 18, 14. [Google Scholar] [CrossRef]
- Bae, D.J.; Jun, J.A.; Chang, H.S.; Park, J.S.; Park, C.S. Epigenetic Changes in Asthma: Role of DNA CpG Methylation. Tubercolosis Respir. Dis. 2020, 83, 1–13. [Google Scholar] [CrossRef]
- Falzone, L.; Salemi, R.; Travali, S.; Scalisi, A.; McCubrey, J.A.; Candido, S.; Libra, M. MMP-9 overexpression is associated with intragenic hypermethylation of MMP9 gene in melanoma. Aging 2016, 8, 933–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeifer, G.P. Defining Driver DNA Methylation Changes in Human Cancer. Int. J. Mol. Sci. 2018, 19, 1166. [Google Scholar] [CrossRef] [Green Version]
- Beacon, T.H.; Delcuve, G.P.; Davie, J.R. Epigenetic regulation of ACE2, the receptor of the SARS-CoV-2 virus(1). Genome 2021, 64, 386–399. [Google Scholar] [CrossRef] [PubMed]
- Tie, Y.; Zhai, C.; Zhang, Y.; Qin, X.; Yu, F.; Li, H.; Shan, M.; Zhang, C. CCAAT/enhancer-binding protein beta overexpression alleviates myocardial remodelling by regulating angiotensin-converting enzyme-2 expression in diabetes. J. Cell Mol. Med. 2018, 22, 1475–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedersen, K.B.; Chodavarapu, H.; Lazartigues, E. Forkhead Box Transcription Factors of the FOXA Class Are Required for Basal Transcription of Angiotensin-Converting Enzyme 2. J. Endocr Soc. 2017, 1, 370–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Demura, M.; Cheng, Y.; Zhu, A.; Karashima, S.; Yoneda, T.; Demura, Y.; Maeda, Y.; Namiki, M.; Ono, K.; et al. Dynamic CCAAT/enhancer binding protein-associated changes of DNA methylation in the angiotensinogen gene. Hypertension 2014, 63, 281–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corley, M.J.; Ndhlovu, L.C. DNA methylation analysis of the COVID-19 host cell receptor, angiotensin I converting enzyme 2 gene (ACE2) in the respiratory system reveal age and gender differences. Preprints 2020, 2020030295. [Google Scholar] [CrossRef] [Green Version]
- Cardenas, A.; Rifas-Shiman, S.L.; Sordillo, J.E.; De Meo, D.L.; Baccarelli, A.A.; Hivert, M.F.; Gold, D.R.; Oken, E. DNA methylation architecture of the ACE2 gene in nasal cells of children. Sci. Rep. 2021, 11, 7107. [Google Scholar] [CrossRef]
- Yang, I.V.; Pedersen, B.S.; Liu, A.H.; O’Connor, G.T.; Pillai, D.; Kattan, M.; Misiak, R.T.; Gruchalla, R.; Szefler, S.J.; Khurana Hershey, G.K.; et al. The nasal methylome and childhood atopic asthma. J. Allergy Clin. Immunol. 2017, 139, 1478–1488. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Yang, J. Colorectal Cancer that Highly Express Both ACE2 and TMPRSS2, Suggesting Severe Symptoms to SARS-CoV-2 Infection. Pathol. Oncol. Res. 2021, 27, 612969. [Google Scholar] [CrossRef]
- Okamoto, Y.; Shinjo, K.; Shimizu, Y.; Sano, T.; Yamao, K.; Gao, W.; Fujii, M.; Osada, H.; Sekido, Y.; Murakami, S.; et al. Hepatitis virus infection affects DNA methylation in mice with humanized livers. Gastroenterology 2014, 146, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Pacis, A.; Tailleux, L.; Morin, A.M.; Lambourne, J.; MacIsaac, J.L.; Yotova, V.; Dumaine, A.; Danckaert, A.; Luca, F.; Grenier, J.C.; et al. Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 2015, 25, 1801–1811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balnis, J.; Madrid, A.; Hogan, K.J.; Drake, L.A.; Chieng, H.C.; Tiwari, A.; Vincent, C.E.; Chopra, A.; Vincent, P.A.; Robek, M.D.; et al. Blood DNA methylation and COVID-19 outcomes. Clin. Epigenet. 2021, 13, 118. [Google Scholar] [CrossRef]
- Liang, W.; Liang, H.; Ou, L.; Chen, B.; Chen, A.; Li, C.; Li, Y.; Guan, W.; Sang, L.; Lu, J.; et al. Development and Validation of a Clinical Risk Score to Predict the Occurrence of Critical Illness in Hospitalized Patients With COVID-19. JAMA Intern. Med. 2020, 180, 1081–1089. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [Green Version]
- Urdinguio, R.G.; Sanchez-Mut, J.V.; Esteller, M. Epigenetic mechanisms in neurological diseases: Genes, syndromes, and therapies. Lancet Neurol. 2009, 8, 1056–1072. [Google Scholar] [CrossRef]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef]
- Pinto, B.G.G.; Oliveira, A.E.R.; Singh, Y.; Jimenez, L.; Goncalves, A.N.A.; Ogava, R.L.T.; Creighton, R.; Schatzmann Peron, J.P.; Nakaya, H.I. ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19. J. Infect. Dis. 2020, 222, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef] [Green Version]
- Clarke, N.E.; Belyaev, N.D.; Lambert, D.W.; Turner, A.J. Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin. Sci. 2014, 126, 507–516. [Google Scholar] [CrossRef]
- Dell’Omo, G.; Crescenti, D.; Vantaggiato, C.; Parravicini, C.; Borroni, A.P.; Rizzi, N.; Garofalo, M.; Pinto, A.; Recordati, C.; Scanziani, E.; et al. Inhibition of SIRT1 deacetylase and p53 activation uncouples the anti-inflammatory and chemopreventive actions of NSAIDs. Br. J. Cancer 2019, 120, 537–546. [Google Scholar] [CrossRef]
- Tikoo, K.; Patel, G.; Kumar, S.; Karpe, P.A.; Sanghavi, M.; Malek, V.; Srinivasan, K. Tissue specific up regulation of ACE2 in rabbit model of atherosclerosis by atorvastatin: Role of epigenetic histone modifications. Biochem. Pharmacol. 2015, 93, 343–351. [Google Scholar] [CrossRef]
- Li, X.; Liu, L.; Yang, S.; Song, N.; Zhou, X.; Gao, J.; Yu, N.; Shan, L.; Wang, Q.; Liang, J.; et al. Histone demethylase KDM5B is a key regulator of genome stability. Proc. Natl. Acad. Sci. USA 2014, 111, 7096–7101. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Cao, J.; Cai, W.L.; Lang, S.M.; Horton, J.R.; Jansen, D.J.; Liu, Z.Z.; Chen, J.F.; Zhang, M.; Mott, B.T.; et al. KDM5 histone demethylases repress immune response via suppression of STING. PLoS Biol. 2018, 16, e2006134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.; Alfajaro, M.M.; DeWeirdt, P.C.; Hanna, R.E.; Lu-Culligan, W.J.; Cai, W.L.; Strine, M.S.; Zhang, S.M.; Graziano, V.R.; Schmitz, C.O.; et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell 2021, 184, 76–91.e13. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Nie, H.; Li, K.; Zhang, Y.X.; Yang, F.; Li, C.B.; Wang, C.F.; Gong, Q. Curcumin inhibits HMGB1 releasing and attenuates concanavalin A-induced hepatitis in mice. Eur. J. Pharmacol. 2012, 697, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Guan, H.; Shi, Q.; Zhang, Y.; Yang, H. Curcumin attenuated acute Propionibacterium acnes-induced liver injury through inhibition of HMGB1 expression in mice. Int. Immunopharmacol. 2015, 24, 159–165. [Google Scholar] [CrossRef]
- Da, W.; Zhang, J.; Zhang, R.; Zhu, J. Curcumin inhibits the lymphangiogenesis of gastric cancer cells by inhibiton of HMGB1/VEGF-D signaling. Int. J. Immunopathol. Pharmacol. 2019, 33, 2058738419861600. [Google Scholar] [CrossRef] [Green Version]
- Cheng, K.; Yang, A.; Hu, X.; Zhu, D.; Liu, K. Curcumin Attenuates Pulmonary Inflammation in Lipopolysaccharide Induced Acute Lung Injury in Neonatal Rat Model by Activating Peroxisome Proliferator-Activated Receptor gamma (PPARgamma) Pathway. Med. Sci. Monit. 2018, 24, 1178–1184. [Google Scholar] [CrossRef]
- Qiao, Y.; Wang, X.M.; Mannan, R.; Pitchiaya, S.; Zhang, Y.; Wotring, J.W.; Xiao, L.; Robinson, D.R.; Wu, Y.M.; Tien, J.C.; et al. Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc. Natl. Acad. Sci. USA 2020. [Google Scholar] [CrossRef]
- Deng, Q.; Rasool, R.U.; Russell, R.M.; Natesan, R.; Asangani, I.A. Targeting androgen regulation of TMPRSS2 and ACE2 as a therapeutic strategy to combat COVID-19. iScience 2021, 24, 102254. [Google Scholar] [CrossRef]
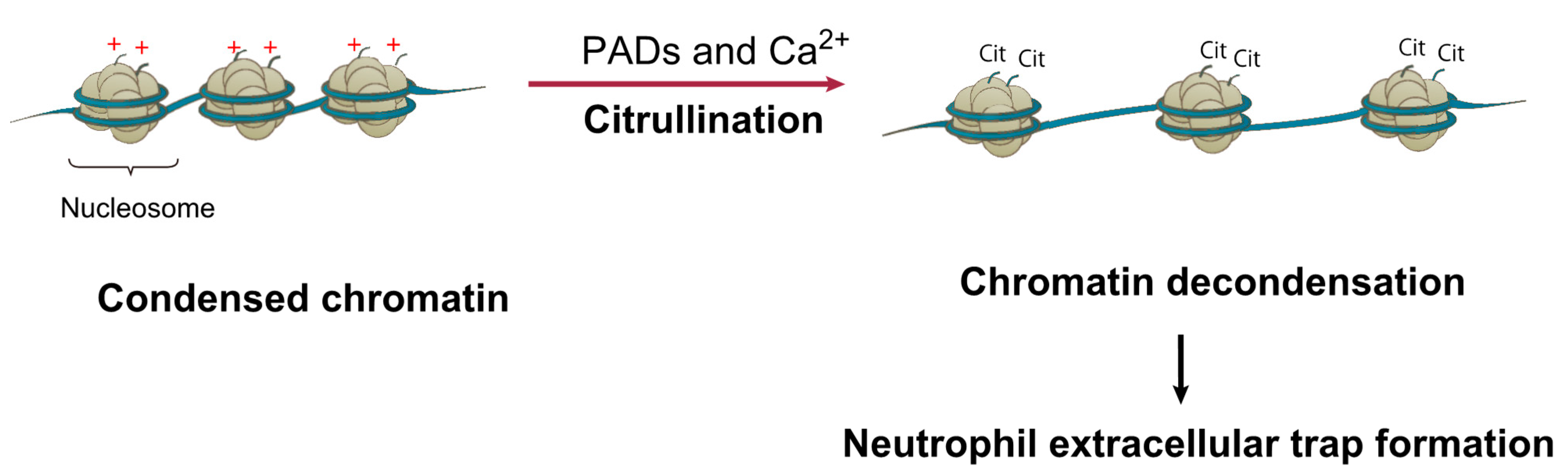
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5, e138999. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wysocka, J.; Sayegh, J.; Lee, Y.H.; Perlin, J.R.; Leonelli, L.; Sonbuchner, L.S.; McDonald, C.H.; Cook, R.G.; Dou, Y.; et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 2004, 306, 279–283. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Stadler, S.; Correll, S.; Li, P.; Wang, D.; Hayama, R.; Leonelli, L.; Han, H.; Grigoryev, S.A.; et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 2009, 184, 205–213. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Li, M.; Lindberg, M.R.; Kennett, M.J.; Xiong, N.; Wang, Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 2010, 207, 1853–1862. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Pfeffer, S.; Zavolan, M.; Grasser, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Johnston, R.J.; Hobert, O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 2003, 426, 845–849. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Vernooy, S.Y.; Guo, M.; Hay, B.A. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003, 13, 790–795. [Google Scholar] [CrossRef] [Green Version]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef] [Green Version]
- Falzone, L.; Grimaldi, M.; Celentano, E.; Augustin, L.S.A.; Libra, M. Identification of Modulated MicroRNAs Associated with Breast Cancer, Diet, and Physical Activity. Cancers 2020, 12, 2555. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, S.; Kenny, A.; Medina, M.; Engel, T.; Jimenez-Mateos, E.M. MicroRNAs in Neurodegenerative Diseases. Int. Rev. Cell Mol. Biol. 2017, 334, 309–343. [Google Scholar] [CrossRef]
- Romaine, S.P.; Tomaszewski, M.; Condorelli, G.; Samani, N.J. MicroRNAs in cardiovascular disease: An introduction for clinicians. Heart 2015, 101, 921–928. [Google Scholar] [CrossRef]
- Garo, L.P.; Murugaiyan, G. Contribution of MicroRNAs to autoimmune diseases. Cell Mol. Life Sci. 2016, 73, 2041–2051. [Google Scholar] [CrossRef] [PubMed]
- Mallick, B.; Ghosh, Z.; Chakrabarti, J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS ONE 2009, 4, e7837. [Google Scholar] [CrossRef] [PubMed]
- Young, D.D.; Connelly, C.M.; Grohmann, C.; Deiters, A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J. Am. Chem. Soc. 2010, 132, 7976–7981. [Google Scholar] [CrossRef]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Chatterjee, S.; Xiao, K.; Riedel, I.; Wang, Y.; Foo, R.; Bar, C.; Thum, T. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J. Mol. Cell Cardiol. 2020, 148, 46–49. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Matarese, A.; Gambardella, J.; Sardu, C.; Santulli, G. miR-98 Regulates TMPRSS2 Expression in Human Endothelial Cells: Key Implications for COVID-19. Biomedicines 2020, 8, 462. [Google Scholar] [CrossRef]
- Vishnubalaji, R.; Shaath, H.; Alajez, N.M. Protein Coding and Long Noncoding RNA (lncRNA) Transcriptional Landscape in SARS-CoV-2 Infected Bronchial Epithelial Cells Highlight a Role for Interferon and Inflammatory Response. Genes 2020, 11, 760. [Google Scholar] [CrossRef]
- Arora, S.; Singh, P.; Dohare, R.; Jha, R.; Ali Syed, M. Unravelling host-pathogen interactions: ceRNA network in SARS-CoV-2 infection (COVID-19). Gene 2020, 762, 145057. [Google Scholar] [CrossRef]
- Mo, Y.; Liu, Y.; Lu, A.; Zhang, H.; Tang, L. Role of circRNAs in viral infection and their significance for diagnosis and treatment (Review). Int. J. Mol. Med. 2021, 47, 88. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Gattuso, G.; Tsatsakis, A.; Spandidos, D.A.; Libra, M. Current and innovative methods for the diagnosis of COVID19 infection (Review). Int. J. Mol. Med. 2021, 47, 100. [Google Scholar] [CrossRef]
- Sood, S.; Aggarwal, V.; Aggarwal, D.; Upadhyay, S.K.; Sak, K.; Tuli, H.S.; Kumar, M.; Kumar, J.; Talwar, S. COVID-19 Pandemic: From Molecular Biology, Pathogenesis, Detection, and Treatment to Global Societal Impact. Curr. Pharmacol. Rep. 2020, 6, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Geng, M.; Peng, Y.; Meng, L.; Lu, S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020, 10, 102–108. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Feng, F.; Hu, G.; Wang, Y.; Yu, Y.; Zhu, Y.; Xu, W.; Cai, X.; Sun, Z.; Han, W.; et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021, 12, 961. [Google Scholar] [CrossRef]
- Daniloski, Z.; Jordan, T.X.; Wessels, H.H.; Hoagland, D.A.; Kasela, S.; Legut, M.; Maniatis, S.; Mimitou, E.P.; Lu, L.; Geller, E.; et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell 2021, 184, 92–105.e116. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.; Fernandez-Justel, J.M.; Santa-Maria, C.; Cadoret, J.C.; Cano-Aroca, L.; Lombrana, R.; Herranz, G.; Agresti, A.; Gomez, M. Chromatin conformation regulates the coordination between DNA replication and transcription. Nat. Commun. 2018, 9, 1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celona, B.; Weiner, A.; Di Felice, F.; Mancuso, F.M.; Cesarini, E.; Rossi, R.L.; Gregory, L.; Baban, D.; Rossetti, G.; Grianti, P.; et al. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol. 2011, 9, e1001086. [Google Scholar] [CrossRef]
- Avgousti, D.C.; Herrmann, C.; Kulej, K.; Pancholi, N.J.; Sekulic, N.; Petrescu, J.; Molden, R.C.; Blumenthal, D.; Paris, A.J.; Reyes, E.D.; et al. A core viral protein binds host nucleosomes to sequester immune danger signals. Nature 2016, 535, 173–177. [Google Scholar] [CrossRef] [Green Version]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.G.; Roberts, C.W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 2011, 11, 481–492. [Google Scholar] [CrossRef]
- Bestle, D.; Heindl, M.R.; Limburg, H.; Van Lam van, T.; Pilgram, O.; Moulton, H.; Stein, D.A.; Hardes, K.; Eickmann, M.; Dolnik, O.; et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 2020, 3. [Google Scholar] [CrossRef]
- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Ashley, E.A. The precision medicine initiative: A new national effort. JAMA 2015, 313, 2119–2120. [Google Scholar] [CrossRef]
- Rosas-Alonso, R.; Colmenarejo-Fernandez, J.; Pernia, O.; Rodriguez-Antolin, C.; Esteban, I.; Ghanem, I.; Sanchez-Cabrero, D.; Losantos-Garcia, I.; Palacios-Zambrano, S.; Moreno-Bueno, G.; et al. Clinical validation of a novel quantitative assay for the detection of MGMT methylation in glioblastoma patients. Clin. Epigenet. 2021, 13, 52. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, F.; Chen, X.; Wang, J.; Zhao, Y.; Li, Y.; He, B. Zinc-dependent Deacetylase (HDAC) Inhibitors with Different Zinc Binding Groups. Curr. Top. Med. Chem. 2019, 19, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.A.; Cahill, K.; Saygin, C.; Odenike, O. Cedazuridine/decitabine: From preclinical to clinical development in myeloid malignancies. Blood Adv. 2021, 5, 2264–2271. [Google Scholar] [CrossRef]
- Hamamoto, R.; Nakamura, Y. Dysregulation of protein methyltransferases in human cancer: An emerging target class for anticancer therapy. Cancer Sci. 2016, 107, 377–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamamoto, R.; Saloura, V.; Nakamura, Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat. Rev. Cancer 2015, 15, 110–124. [Google Scholar] [CrossRef]
- Rugo, H.S.; Jacobs, I.; Sharma, S.; Scappaticci, F.; Paul, T.A.; Jensen-Pergakes, K.; Malouf, G.G. The Promise for Histone Methyltransferase Inhibitors for Epigenetic Therapy in Clinical Oncology: A Narrative Review. Adv. Ther. 2020, 37, 3059–3082. [Google Scholar] [CrossRef]
- Hillyar, C.; Rallis, K.S.; Varghese, J. Advances in Epigenetic Cancer Therapeutics. Cureus 2020, 12, e11725. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaneko, S.; Takasawa, K.; Asada, K.; Shinkai, N.; Bolatkan, A.; Yamada, M.; Takahashi, S.; Machino, H.; Kobayashi, K.; Komatsu, M.; et al. Epigenetic Mechanisms Underlying COVID-19 Pathogenesis. Biomedicines 2021, 9, 1142. https://doi.org/10.3390/biomedicines9091142
Kaneko S, Takasawa K, Asada K, Shinkai N, Bolatkan A, Yamada M, Takahashi S, Machino H, Kobayashi K, Komatsu M, et al. Epigenetic Mechanisms Underlying COVID-19 Pathogenesis. Biomedicines. 2021; 9(9):1142. https://doi.org/10.3390/biomedicines9091142
Chicago/Turabian StyleKaneko, Syuzo, Ken Takasawa, Ken Asada, Norio Shinkai, Amina Bolatkan, Masayoshi Yamada, Satoshi Takahashi, Hidenori Machino, Kazuma Kobayashi, Masaaki Komatsu, and et al. 2021. "Epigenetic Mechanisms Underlying COVID-19 Pathogenesis" Biomedicines 9, no. 9: 1142. https://doi.org/10.3390/biomedicines9091142
APA StyleKaneko, S., Takasawa, K., Asada, K., Shinkai, N., Bolatkan, A., Yamada, M., Takahashi, S., Machino, H., Kobayashi, K., Komatsu, M., & Hamamoto, R. (2021). Epigenetic Mechanisms Underlying COVID-19 Pathogenesis. Biomedicines, 9(9), 1142. https://doi.org/10.3390/biomedicines9091142






