Secreted Trimeric Chikungunya Virus Spikes from Insect Cells: Production, Purification, and Glycosylation Status
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Recombinant Protein Production and Purification
2.3. Furin Cleavage Site Analysis
2.4. PNGase F Treatment
2.5. Bis(Sulfosuccinimidyl)Suberate Cross-Linking
2.6. Western Blotting
2.7. LC–MS/MS-Based Glycoproteomic Analysis
3. Results
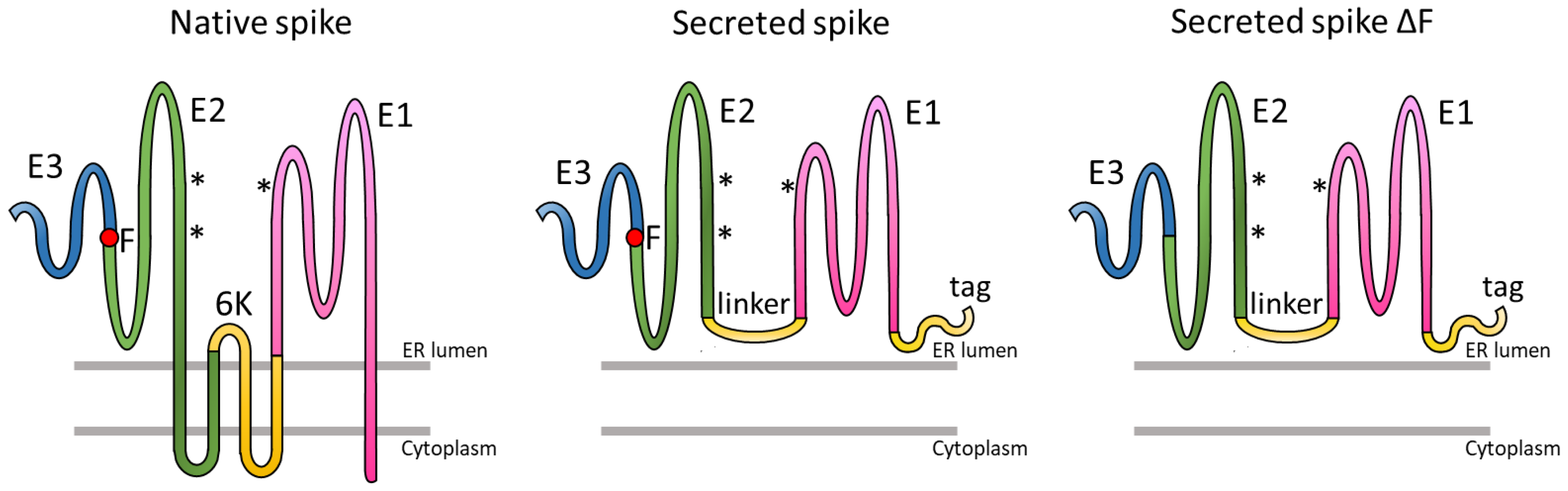
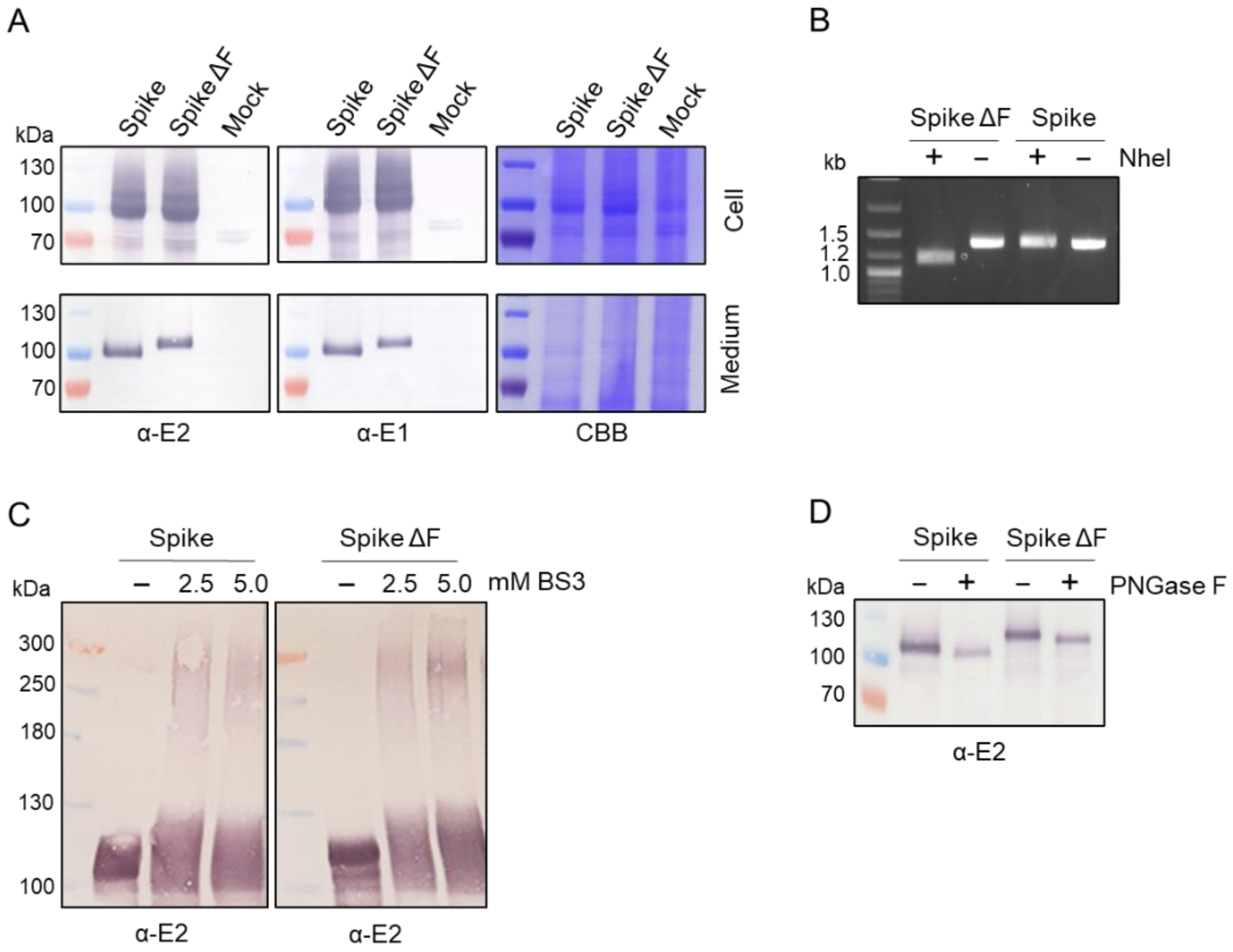
3.1. Expression of Trimeric, Glycosylated Chikungunya Virus Spikes in Insect Cells
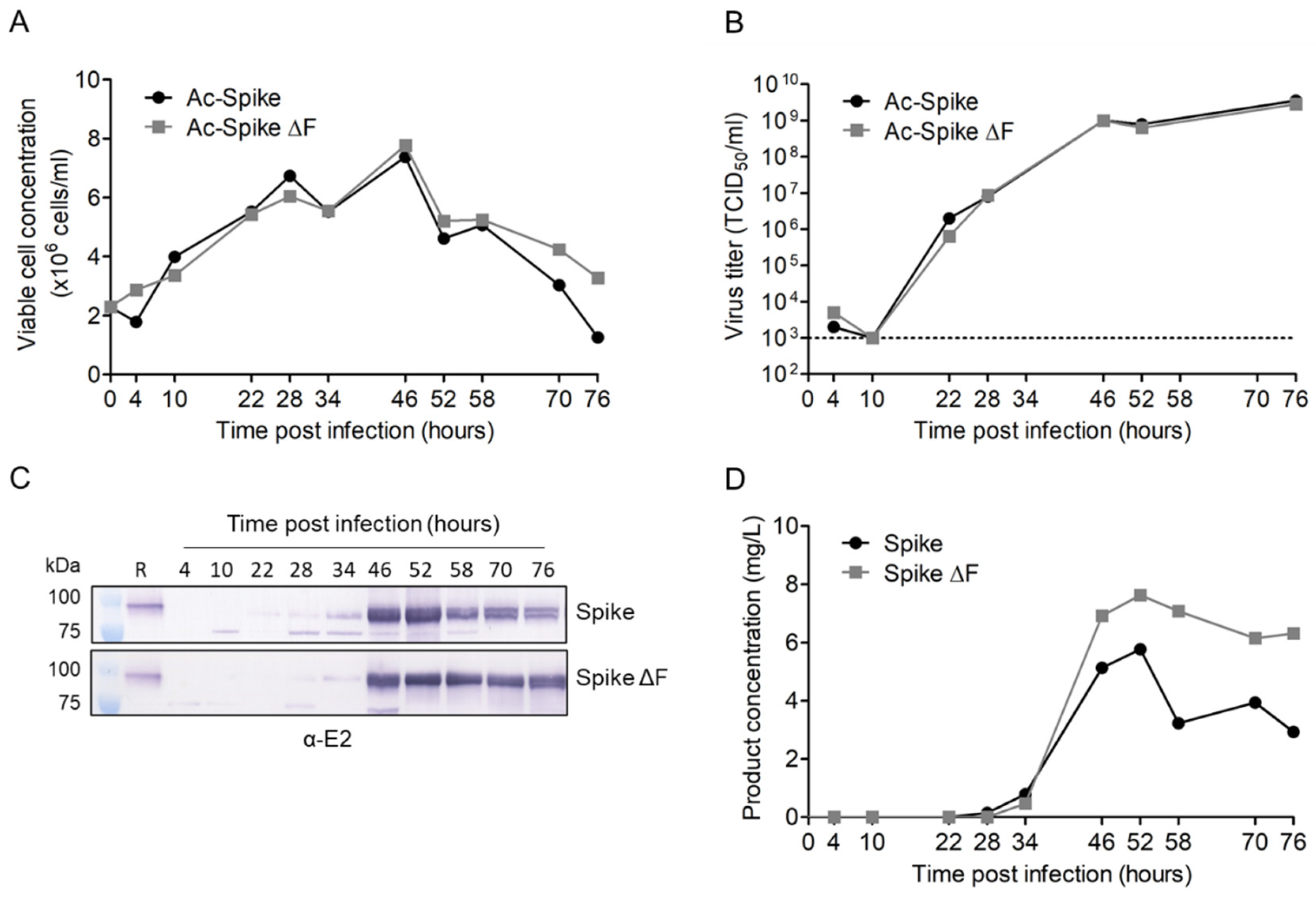
3.2. Production of Secreted CHIKV Spikes in Sf9 Suspension Cells
3.3. Downstream Processing by Strep-Tactin Affinity Column Purification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Borgherini, G.; Poubeau, P.; Jossaume, A.; Gouix, A.; Cotte, L.; Michault, A.; Arvin-Berod, C.; Paganin, F. Persistent Arthralgia Associated with Chikungunya Virus: A Study of 88 Adult Patients on Reunion Island. Clin. Infect. Dis. 2008, 47, 469–475. [Google Scholar] [CrossRef]
- Wahid, B.; Ali, A.; Rafique, S.; Idrees, M. Global expansion of chikungunya virus: Mapping the 64-year history. Int. J. Infect. Dis. 2017, 58, 69–76. [Google Scholar] [CrossRef]
- Gao, S.; Song, S.; Zhang, L. Recent Progress in Vaccine Development Against Chikungunya Virus. Front. Microbiol. 2019, 10, 2881. [Google Scholar] [CrossRef]
- Schrauf, S.; Tschismarov, R.; Tauber, E.; Ramsauer, K. Current Efforts in the Development of Vaccines for the Prevention of Zika and Chikungunya Virus Infections. Front. Immunol. 2020, 11, 592. [Google Scholar] [CrossRef]
- Van Oers, M.M.; Pijlman, G.P.; Vlak, J.M. Thirty years of baculovirus-insect cell protein expression: From dark horse to mainstream technology. J. Gen. Virol. 2015, 96, 6–23. [Google Scholar] [CrossRef]
- Chen, N.; Kong, X.; Zhao, S.; Xiaofeng, W. Post-translational modification of baculovirus-encoded proteins. Virus Res. 2020, 279, 197865. [Google Scholar] [CrossRef]
- van Oers, M.M. Opportunities and challenges for the baculovirus expression system. J. Invertebr. Pathol. 2011, 107, S3–S15. [Google Scholar] [CrossRef]
- Pijlman, G.P.; Grose, C.; Hick, T.A.H.; Breukink, H.E.; Braak, R.V.D.; Abbo, S.R.; Geertsema, C.; Van Oers, M.M.; Martens, D.E.; Esposito, D. Relocation of the attTn7 Transgene Insertion Site in Bacmid DNA Enhances Baculovirus Genome Stability and Recombinant Protein Expression in Insect Cells. Viruses 2020, 12, 1448. [Google Scholar] [CrossRef]
- Sun, B.; Zhao, D.; Zhang, X.; Gu, T.; Yu, X.; Sun, S.; Zhao, X.; Wei, L.; Liu, D.; Yan, H.; et al. Development a scalable production process for truncated human papillomavirus type-6 L1 protein using WAVE Bioreactor and hollow fiber membrane. Appl. Microbiol. Biotechnol. 2015, 100, 1231–1240. [Google Scholar] [CrossRef]
- Wagner, J.M.; Pajerowski, J.D.; Daniels, C.L.; McHugh, P.M.; Flynn, J.A.; Balliet, J.W.; Casimiro, D.R.; Subramanian, S. Enhanced production of chikungunya virus-like particles using a high-pH adapted Spodoptera frugiperda insect cell line. PLoS ONE 2014, 9, e94401. [Google Scholar]
- Weber, W.; Weber, E.; Geisse, S.; Memmert, K. Optimisation of protein expression and establishment of the Wave Bioreactor for Baculovirus/insect cell culture. Cytotechnology 2002, 38, 77–85. [Google Scholar] [CrossRef]
- Metz, S.W.; Gardner, J.; Geertsema, C.; Le, T.T.; Goh, L.; Vlak, J.M.; Suhrbier, A.; Pijlman, G.P. Effective Chikungunya Virus-like Particle Vaccine Produced in Insect Cells. PLoS Neglected Trop. Dis. 2013, 7, e2124. [Google Scholar] [CrossRef] [PubMed]
- Metz, S.W.; Martina, B.E.; Doel, P.V.D.; Geertsema, C.; Osterhaus, A.; Vlak, J.M.; Pijlman, G. Chikungunya virus-like particles are more immunogenic in a lethal AG129 mouse model compared to glycoprotein E1 or E2 subunits. Vaccine 2013, 31, 6092–6096. [Google Scholar] [CrossRef]
- Voss, J.E.; Vaney, M.-C.; Duquerroy, S.; Vonrhein, C.; Girard-Blanc, C.; Crublet, E.; Thompson, A.; Bricogne, G.; Rey, F. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 2010, 468, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, R.J. Togaviridae: The viruses and their replication. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams and Wilkins: Baltimore, MD, USA, 2007; pp. 1001–1022. [Google Scholar]
- Mulvey, M.; Brown, D.T. Assembly of the Sindbis Virus Spike Protein Complex. Virology 1996, 219, 125–132. [Google Scholar] [CrossRef]
- Zhang, X.; Fugère, M.; Day, R.; Kielian, M. Furin Processing and Proteolytic Activation of Semliki Forest Virus. J. Virol. 2003, 77, 2981–2989. [Google Scholar] [CrossRef]
- Sjöberg, M.; Lindqvist, B.; Garoff, H. Activation of the alphavirus spike protein is suppressed by bound E3. J. Virol. 2011, 85, 5644–5650. [Google Scholar] [CrossRef]
- Yap, M.L.; Klose, T.; Urakami, A.; Hasan, S.S.; Akahata, W.; Rossmann, M.G. Structural studies of Chikungunya virus maturation. Proc. Natl. Acad. Sci. USA 2017, 114, 13703–13707. [Google Scholar] [CrossRef]
- Hopkins, R.F.; Esposito, D. A rapid method for titrating baculovirus stocks using the Sf-9 Easy Titer cell line. BioTechniques 2009, 47, 785–788. [Google Scholar] [CrossRef]
- Urakami, A.; Sakurai, A.; Ishikawa, M.; Yap, M.L.; Flores-Garcia, Y.; Haseda, Y.; Aoshi, T.; Zavala, F.P.; Rossmann, M.G.; Kuno, S.; et al. Development of a Novel Virus-Like Particle Vaccine Platform That Mimics the Immature Form of Alphavirus. Clin. Vaccine Immunol. 2017, 24, e00090-17. [Google Scholar] [CrossRef]
- Akahata, W.; Nabel, G.J. A Specific Domain of the Chikungunya Virus E2 Protein Regulates Particle Formation in Human Cells: Implications for Alphavirus Vaccine Design. J. Virol. 2012, 86, 8879–8883. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jose, J.; Xiang, Y.; Kuhn, R.J.; Rossmann, M.G. Structural changes of envelope proteins during alphavirus fusion. Nature 2010, 468, 705–708. [Google Scholar] [CrossRef]
- Akahata, W.; Yang, Z.-Y.; Andersen, H.; Sun, S.; Holdaway, H.A.; Kong, W.-P.; Lewis, M.G.; Higgs, S.; Rossmann, M.G.; Rao, S.; et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat. Med. 2010, 16, 334–338. [Google Scholar] [CrossRef]
- Van Oosten, L.; Altenburg, J.J.; Fougeroux, C.; Geertsema, C.; Den End, F.; Evers, W.A.C.; Westphal, A.H.; Lindhoud, S.; van den Berg, W.; Swarts, D.C.; et al. Two-component nanoparticle vaccine displaying glycosylated spike S1 domain induces neutralizing antibody response against SARS-CoV-2. MBio 2021, 12, e01813-21. [Google Scholar] [CrossRef] [PubMed]
- Buckland, B.; Boulanger, R.; Fino, M.; Srivastava, I.; Holtz, K.; Khramtsov, N.; McPherson, C.; Meghrous, J.; Kubera, P.; Cox, M.M. Technology transfer and scale-up of the Flublok® recombinant hemagglutinin (HA) influenza vaccine manufacturing process. Vaccine 2014, 32, 5496–5502. [Google Scholar] [CrossRef]
- Buckland, B.C. The development and manufacture of influenza vaccines. Hum. Vaccin. Immunother. 2015, 11, 1357–1360. [Google Scholar] [CrossRef]
- Vicente, T.; Roldao, A.; Peixoto, C.; Carrondo, M.J.; Alves, P. Large-scale production and purification of VLP-based vaccines. J. Invertebr. Pathol. 2011, 107, S42–S48. [Google Scholar] [CrossRef]
- Krammer, F.; Schinko, T.; Palmberger, D.; Tauer, C.; Messner, P.; Grabherr, R. Trichoplusia ni cells (High FiveTM) are highly efficient for the production of influenza A virus-like particles: A comparison of two insect cell lines as production platforms for influenza vaccines. Mol. Biotechnol. 2010, 45, 226–234. [Google Scholar] [CrossRef]
- Ikonomou, L.; Schneider, Y.-J.; Agathos, S.N. Insect cell culture for industrial production of recombinant proteins. Appl. Microbiol. Biotechnol. 2003, 62, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, F.; Bernal, V.; Chaillet, M.; Berger, I.; Alves, P.M. Targeted supplementation design for improved production and quality of enveloped viral particles in insect cell-baculovirus expression system. J. Biotechnol. 2016, 233, 34–41. [Google Scholar] [CrossRef][Green Version]
- Carinhas, N.; Bernal, V.; Monteiro, F.; Carrondo, M.; Oliveira, R.; Alves, P. Improving baculovirus production at high cell density through manipulation of energy metabolism. Metab. Eng. 2010, 12, 39–52. [Google Scholar] [CrossRef]
- Marek, M.; van Oers, M.M.; Devaraj, F.F.; Vlak, J.M.; Merten, O.-W. Engineering of baculovirus vectors for the manufacture of virion-free biopharmaceuticals. Biotechnol. Bioeng. 2010, 108, 1056–1067. [Google Scholar] [CrossRef]
- Chen, G.L.; Coates, E.E.; Plummer, S.H.; Carter, C.A.; Berkowitz, N.; Conan-Cibotti, M.; Cox, J.H.; Beck, A.; O’Callahan, M.; Andrews, C.; et al. Effect of a chikungunya virus-like particle vaccine on safety and tolerability outcomes: A randomized clinical trial. J. Am. Med. Assoc. 2020, 323, 1369–1377. [Google Scholar] [CrossRef]
- Chang, L.-J.; Dowd, K.A.; Mendoza, F.H.; Saunders, J.G.; Sitar, S.; Plummer, S.H.; Yamshchikov, G.; Sarwar, U.N.; Hu, Z.; Enama, M.E.; et al. Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: A phase 1 dose-escalation trial. Lancet 2014, 384, 2046–2052. [Google Scholar] [CrossRef]
- Brandler, S.; Ruffié, C.; Combredet, C.; Brault, J.-B.; Najburg, V.; Prevost, M.-C.; Habel, A.; Tauber, E.; Desprès, P.; Tangy, F. A recombinant measles vaccine expressing chikungunya virus-like particles is strongly immunogenic and protects mice from lethal challenge with chikungunya virus. Vaccine 2013, 31, 3718–3725. [Google Scholar] [CrossRef]
- Arévalo, M.T.; Huang, Y.; Jones, C.A.; Ross, T.M. Vaccination with a chikungunya virus-like particle vaccine exacerbates disease in aged mice. PLoS Neglected Trop. Dis. 2019, 13, e0007316. [Google Scholar] [CrossRef]
- Garg, H.; Mehmetoglu-Gurbuz, T.; Joshi, A. Virus Like Particles (VLP) as multivalent vaccine candidate against Chikungunya, Japanese Encephalitis, Yellow Fever and Zika Virus. Sci. Rep. 2020, 10, 4017. [Google Scholar] [CrossRef]
- García-Arriaza, J.; Cepeda, V.; Hallengärd, D.; Sorzano, C.Ó.S.; Kümmerer, B.M.; Liljeström, P.; Esteban, M. A novel poxvirus-based vaccine, MVA-CHIKV, is highly immunogenic and protects mice against chikungunya infection. J. Virol. 2014, 88, 3527–3547. [Google Scholar] [CrossRef]
- Fong, R.H.; Banik, S.S.R.; Mattia, K.; Barnes, T.; Tucker, D.; Liss, N.; Lu, K.; Selvarajah, S.; Srinivasan, S.; Mabila, M.; et al. Exposure of Epitope Residues on the Outer Face of the Chikungunya Virus Envelope Trimer Determines Antibody Neutralizing Efficacy. J. Virol. 2014, 88, 14364–14379. [Google Scholar] [CrossRef]
- Dora, E.G.; Rossi, S.L.; Weaver, S.C.; Tucker, S.N.; Mateo, R. An adjuvanted adenovirus 5-based vaccine elicits neutralizing antibodies and protects mice against chikungunya virus-induced footpad swelling. Vaccine 2019, 37, 3146–3150. [Google Scholar] [CrossRef]
- López-Camacho, C.; Kim, Y.C.; Blight, J.; Moreli, M.L.; Montoya-Diaz, E.; Huiskonen, J.T.; Kümmerer, B.M.; Reyes-Sandoval, A. Assessment of Immunogenicity and Neutralisation Efficacy of Viral-Vectored Vaccines Against Chikungunya Virus. Viruses 2019, 11, 322. [Google Scholar] [CrossRef]
- Zhang, B.; Chao, C.W.; Tsybovsky, Y.; Abiona, O.M.; Hutchinson, G.B.; Moliva, J.I.; Olia, A.S.; Pegu, A.; Phung, E.; Stewart-Jones, G.B.E.; et al. A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone. Sci. Rep. 2020, 10, 18149. [Google Scholar] [CrossRef]
- Wichgers Schreur, P.J.; Tacken, M.; Gutjahr, B.; Keller, M.; van Keulen, L.; Kant, J.; van de Water, S.; Lin, Y.; Eiden, M.; Rissmann, M.; et al. Vaccine efficacy of self-assembled multimeric protein scaffold particles displaying the glycoprotein Gn head domain of rift valley fever virus. Vaccines 2021, 9, 301. [Google Scholar] [CrossRef]
- Fougeroux, C.; Goksøyr, L.; Idorn, M.; Soroka, V.; Myeni, S.K.; Dagil, R.; Janitzek, C.M.; Søgaard, M.; Aves, K.-L.; Horsted, E.W.; et al. Capsid-like particles decorated with the SARS-CoV-2 receptor-binding domain elicit strong virus neutralization activity. Nat. Commun. 2021, 12, 324. [Google Scholar] [CrossRef]
- Kam, Y.-W.; Lee, W.W.L.; Simarmata, D.; Harjanto, S.; Teng, T.-S.; Tolou, H.; Chow, A.; Lin, R.T.P.; Leo, Y.-S.; Rénia, L.; et al. Longitudinal Analysis of the Human Antibody Response to Chikungunya Virus Infection: Implications for Serodiagnosis and Vaccine Development. J. Virol. 2012, 86, 13005–13015. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hick, T.A.H.; Geertsema, C.; Henquet, M.G.L.; Martens, D.E.; Metz, S.W.; Pijlman, G.P. Secreted Trimeric Chikungunya Virus Spikes from Insect Cells: Production, Purification, and Glycosylation Status. Processes 2022, 10, 162. https://doi.org/10.3390/pr10010162
Hick TAH, Geertsema C, Henquet MGL, Martens DE, Metz SW, Pijlman GP. Secreted Trimeric Chikungunya Virus Spikes from Insect Cells: Production, Purification, and Glycosylation Status. Processes. 2022; 10(1):162. https://doi.org/10.3390/pr10010162
Chicago/Turabian StyleHick, Tessy A. H., Corinne Geertsema, Maurice G. L. Henquet, Dirk E. Martens, Stefan W. Metz, and Gorben P. Pijlman. 2022. "Secreted Trimeric Chikungunya Virus Spikes from Insect Cells: Production, Purification, and Glycosylation Status" Processes 10, no. 1: 162. https://doi.org/10.3390/pr10010162
APA StyleHick, T. A. H., Geertsema, C., Henquet, M. G. L., Martens, D. E., Metz, S. W., & Pijlman, G. P. (2022). Secreted Trimeric Chikungunya Virus Spikes from Insect Cells: Production, Purification, and Glycosylation Status. Processes, 10(1), 162. https://doi.org/10.3390/pr10010162







