Abstract
Trastuzumab is a target-based recombinant humanized IgG1 monoclonal antibody (mAbs), extensively employed for treatment of metastatic breast cancer with human epidermal growth receptor 2 (HER2) overexpression. Studies around the world have reported that mAbs have substantial inter-patient unpredictable absorption, distribution, metabolism, and excretion (ADME-pharmacokinetics) because of multiple elements manipulating the concentration of mAbs in plasma. Herein, we have established a bioanalytical technique using UPLC-MS/MS with an easy sample workup method and in-solution digestion protocol to assay the trastuzumab plasma samples from breast cancer patients in clinical studies. Surrogated proteolytic peptides were used for accurate quantification of trastuzumab (CanMab) with a trastuzumab signature peptide with [13C6, 15N4]-arginine and [13C6, 15N2]-lysine stable isotope-labeled (SIL) peptide. Experiments to validate the method were accurately carried out according to the guidelines mentioned in the bioanalytical method validation protocol. The evaluation established excellent linearity over a wide range of 5–500 µg/mL. The experimental procedure was efficaciously performed in a pilot study of five breast cancer patients and residual concentrations of drugs from responding and non-responding subjects were compared. The receiver operating characteristic (ROC) examination displayed that 52.25 µg/mL was the Cmin threshold predictive response with a satisfactory sensitivity of 88.58% and specificity of 79.25%.
1. Introduction
HER2 overexpressing metastatic breast and gastric cancer are mostly diagnosed cancers across the world. These cancers have overexpressed cell surface HER2 receptors, and it is proposed that the overexpression or gene amplification of HER2 has been traced in about 20–30% of breast cancers [1]. Elevated activation of HER2 results in multiple downstream pathways leading to unusual ontogenesis of cancerous cells. The over-expression of HER2 in breast cancer cells triggers the amplification of the signal transduced by other receptors of the HER family via the formation of homodimers and heterodimers. The HER2 receptor, also called the ‘transmembrane tyrosine kinase receptor’ is comprised of a ligand-binding domain which is extracellular, intracellular, or cytoplasmic tyrosine kinase domain and a transmembrane domain. The activation of HER2 commences with the formation of homodimers or heterodimers with other EGFR proteins, resulting in dimerization and autophosphorylation and/or transphosphorylation of particular tyrosine kinase residue in EGFR domains which are intracellular in nature. The downstream molecular cascade is further activated, such as Ras/Raf/nitrogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase/Akt, and the phospholipase Cγ (PLCγ)/protein kinase C (PKC) pathways that promote cell growth and survival and cell cycle progression and block cell cycle advancement by prompting the establishment of p27/Cdk2 centers/complexes. Abnormal cell proliferation and hyper activation of these signaling pathways are observed in cancer cells due to the upregulation of HER2 [2,3].
Trastuzumab is an IgG1 Kappa humanized monoclonal antibody (MAB), derived from Chinese hamster ovary (CHO) cell culture, and it is widely used to treat various malignancies, including adjuvant or metastatic breast cancer and gastric cancer. Trastuzumab has been found to mediate the activation of antibody-dependent cell-mediated cytotoxicity(ADDC) by attracting immune cells, such as natural killer cells, to the tumor sites that overexpress HER2 receptors and this is the reason why it is primarily used for the HER positive breast cancer treatment [4]. Trastuzumab recieved initial USFDA approval in 1998 (HERCLON) for the treatment of cancer [5]. Now the exclusive right of the HERCLON has expired, and several biosimilars of trastuzumab, including OGIVRI (trastuzumab-dkst), HERZUMA (trastuzumab-pkrb), and KANJINTI (trastuzumab-anns) have been accepted for the management of breast and gastric cancers with overexpressed HER2. The recommended dose for trastuzumab administered as intravenous (IV) infusions ranged between 10 and 500 mg once in a week with a starting preliminary 8 mg/kg dose which is permeated over a period of 90 min, which was later decreased to 6 mg/kg over a period of 30 to 90 min every 3 weeks. Trastuzumab has a dose-dependent pharmacokinetic profile, with an average t1/2 between 2 and 12 days at the recommended dose of 10–500 mg [6,7]. At maximum dose of 500 mg, the plasma concentrations (Cmax) were found to be 377 µg/mL [8].
In the case of mAbs, a prominent interpatient variability has been observed. The interpatient variability in clearance and distribution (43%, 29%) has been studied and reported for trastuzumab. The clearance of mAbs is inversely proportional to the minimum effective concentration, resulting in less trough concentrations with increased clearance, leading to significant illness management at completion of the dosing period. Therefore, we must understand the mechanisms governing interpatient variability in mAbs pharmacokinetics to get insight into how to achieve optimal clinical response. It has been reported that exposure–response data displayed that subjects with decreased plasma trough concentrations are at the risk of failure of treatment protocol. Subjects reflecting maximum diseased state can be seized in a deprived exposure–response phase and are the ones who need optimization of doses for better pharmacological response [9].
TDM helps in monitoring the therapeutic effectiveness of drug molecules and assists in correlating the loss of response of mAbs. Here, we have employed triple-quadrupole (TQD) mass spectrometer that can be employed for LC-MS and LC-MS/MS methods. Preparation of samples includes: (a) the separation of heavy chains from lighter chains after reducing disulfide bonds; and (b) cleavage using trypsin to produce numerous peptide chains from the immunoglobulin. For quantitative analysis of mAbs, peptides which are particular to the flexible section of mAbs on either light or heavy chains and these regions did not undergo cross-reaction with human sequences. In order to mollify the peptide load, plasma samples of mAbs are either demodulated or supplemented. The extracted/demodulated samples were then denatured using guanidine HCL (unfolding of protein), reduced using dithiothreitol (reduction of cysteine residue causes breakdown of disulfide bonds linking light and heavy chains of mAb), alkylated using iodoacetamide (cysteine residues alkylation inhibits reformation of disulfide bonds), and finally processed by trypsin into reduced fragments of peptide chain. This mixture of peptide must be parted using liquid chromatography before loading the sample for mass spectrometry (MS/MS). After the separation is completed, the mass of the interested peptide is recorded. In the mass spectrometer, the peptide chain is cleaved/delinked forming an ion fragment, and the transition of ion pair which is employed for quantification, on the TQD [10].
We have employed sequencing grade trypsin to digest the mAbs and to achieve effective digestion of proteins which is a critical step in producing targeted peptides required for perfect quantification. Unavailability of signature peptides necessary for perfect quantification is attributed to incomplete digestion of peptides. To overcome this, we can incorporate the trypsin cleavage in the synthetic internal standard for protein quantification. The good thing with trypsin is that it is highly specific, assertive, stable, and cleaves exclusively C-terminal region to arginine residues and lysine residues. The advantage of cleaving C-terminal region to arginine residues or lysine residues results in peptide generation in the desired mass range for fragmentation through tandem-mass spectrometry (MS/MS) and exceedingly basic residues are then placed at the C-terminal region of the peptides. This leads to generation of high mass y-ion sequences and enables tandem mass spectrometry to be effortlessly accountable [11].
In protein bio-therapeutics including mAbs, the surrogate peptide methodology is extensively employed for quantification. It is of the right choice to use surrogate peptides employing a TQM spectrometer because the unique selectivity of the instrument is very efficient in distinguishing the same element of isotopes of endogenous peptides and surrogate characterized peptides. The high selectivity and specificity of the instrument precisely correct the variability that may occur due to essential properties of peptides and undistinguishable separation data obtained through chromatography. MS ionization offers reliable quantification and facilitates the determination of absolute richness. The threats that are linked with background signal interferences originating from intricate natural matrices are alleviated using a surrogate peptide approach. This approach offers accurate and highly reliable quantification of huge biomolecules and does not require development of particular antibodies [12,13].
Here, we have employed the LC-MS/MS approach to demonstrate the implementation of TDM strategies at the bedside. The variation in the PK parameters (clearance) that is associated with drug exposure levels demands such techniques that have the potential to forecast the drug efficacy and adapt dosing when the drug exposure levels in the patients are insufficient. The employment of TDM in routine clinical exercise requires a rapid and roust bioanalytical method that will ensure the applicability of the method in real clinical settings [14]. Operating LC-MS/MS is very interesting and challenging as well because of intense multiplicity and density of plasma proteins [15]. To overcome this challenge, we have employed efficient preparation of samples to eliminate possible interferences. The method developed offers an easy sample preparation workflow, meeting the routine requirement in relations to cost, efficiency, and analysis. This pilot study is designed to reflect a systematic demonstration of original procedure and to assess whether easy and quick management of remaining and/or maximal plasma concentrations of trastuzumab could be an admissible approach to anticipate management effectiveness in breast cancer patients. The developed assay was successfully employed in a pilot study of 20 breast cancer patients and residual concentrations of drugs from responding and non-responding subjects were compared.
2. Materials and Methods
2.1. Chemicals and Materials
Trastuzumab (CANMAb) as a standard drug (150 mg) was obtained from Biocon (Mumbai, India). Acetonitrile (LC-MS grade), methanol, dithiothreitol, and iodoacetamide were procured from Sigma Aldrich (St. Louis, MO, USA). Formic acid and ammonium acetate were obtained from Fluka Chemical (Darmstadt, Germany). Sequencing grade trypsin was procured from Promega (Madison, WI, USA). Internal standard (ISTD) of trastuzumab signature peptide with a heavy labeled [13C6, 15N4]-arginine and [13C6, 15N2]-lysine was acquired from Sigma Aldrich (St. Louis, MO, USA).
2.2. Preparation of Standard Solution
Stock solutions of standard drug trastuzumab were prepared at a concentration of 15 mg/mL in bacteriostatic water for injection (BWFI) and kept at 4 °C. Intermediate spiking solutions at a concentration of 2 mg/mL were obtained employing fresh human plasma. Serially diluted intermediate solution with blank plasma to obtain standards (5, 10, 20, 50, 100, 200, 400, and 500 µg/mL) and QC samples (15.2, 249, and 396 µg/mL). Stock solution of internal standard trastuzumab (100.0 µg/mL) was obtained at 1.0 mg/mL concentration. All CC and QC samples and internal standard solutions were prepared on ice and finally stored at −65 ± 10 °C.
2.3. Preparation of Sample
A simple and fast workflow was developed and aimed at routine patient management of trastuzumab concentration (Figure 1). In solution, tryptic digestion was performed using standard protocol. A quantity of 400 µL from 2 mg/mL (drug concentration) was denatured using guanidine HCL (6 M, 60 µL) in 20 mM Tris HCL (200 µL). The solution obtained was further processed for disulfide bond reduction using 60 µL of 20 mM DTT (dithiothreitol) at 60 °C for 2 h. It was then alkylated, employing 170 µL of 20 mM IAMI iodoacetamide at 25 °C in the dark. Sequencing grade modified trypsin (15 µg/sample) and a sample (mixed together) were incubated at 37 °C whole night for tryptic digestion. The tryptic digest plasma sample was appeased/quenched using 0.1% formic acid.
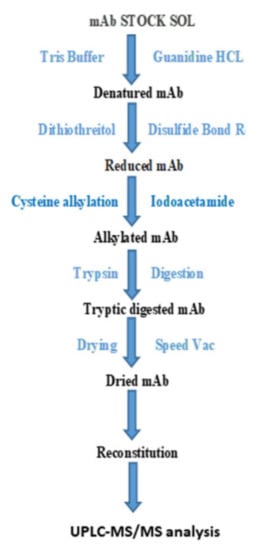
Figure 1.
Graphical description of peptide digestion workflow for trastuzumab analysis.
Desalinization of acidified tryptic peptide mixture was carried out using SUPEL-SELECT HLB (30 mg/L mL) SPE cartridges. The cartridges were maintained with methanol (1 mL, LC-MS grade) and formic acid solution (1 mL, 0.1%) on solid-phase extraction multiplies by employing low pressure. Acidified tryptic digest mixture was loaded on cartridges and washed away with a formic acid solution (1 mL, 0.1%). Peptides were then eluted using 30 µL (2×) of elution mixture; 2% NH4OH in acetonitrile and water (50:50) into pre-labeled gathering tubes. Eluted samples were speed Vac concentrated under ambient temperature conditions. The dried tryptic peptide mixture was reconstructed in formic acid (30 mL, 0.1%) and acetonitrile (5%) for analysis using UPLC-MS/MS.
2.4. Mass Spectrometry and Chromatographic Conditions
Mass spectrometric analyses were carried out using UPLC-MS/MS on XEVO TQ-S triple quadrupole mass spectrometer (Waters, Milford, MA, USA) operated using MassLynx software (version 4.1). The spectrometer was connected to an Acquity I-Class UPLC system (Waters, Milford, MA, USA). Chromatographic separation was carried out employing a gradient combination of solvent A (0.1% solution of HCOOH in water) and B (0.1% HCOOH in acetonitrile). Peptide chains were parted on an Acquity UPLC BEH Shield C18 column, 2.1 × 50 mm, 1.7 μm, (Waters, Milford, MA, USA) using a gradient B (5 to 33%) for a period of 10 min along with a flow rate of 0.3 mL/min at 40 °C. MS/MS data were obtained using electrospray ionization technique in positive mode and multiple reaction monitoring mode (MRM) was employed for the transition of selected peptides using a source, ion spray voltage of 3000 V, and a desolvation temperature ranging between 150 and 450 °C, respectively. The entrance potential was kept at 12 eV and cone voltage was maintained at 40 V.
2.5. Assay Validation
The devolved assay was validated as per guidelines cited by USFDA for validation of bioanalytical method [16]. The validated factors/parameters include linearity, selectivity, matrix effects, and correctness. Recognition measures for calibration standards (±20%) and QCs (±25% at the LLOQ) were carried out in line as per recommendations for LC-MS/MS methods [17].
2.6. Patients
A total of 20 breast cancer patients (all females), mean age (68), and range (48–81) were enrolled for this pilot study undergoing treatment at Batra Hospital and Research Centre New Delhi. All the patients were managed with trastuzumab comprising treatment regimen from October 2018 to May 2019. Six patients/subjects were earlier treated with radiotherapy and 11 patients were managed using at least one line of chemotherapy (combination X). All the patients were informed about the study and informed consent was signed prior to sampling. The approval from local ethics committee (Scientific Research and Ethics Review Committee Batra Hospital and Research Centre 1, Tughlaqabad Institutional Area M.B. Road, New Delhi-110062) with Letter No. SRERC/2018/19/09 and Submission No. SRERC/2018/TH/24 was obtained before the commencement of the study. Throughout the whole study, all the experiments were performed according to the latest ethical guidelines. Sampling of blood was carried out by skilled nurses using standard protocol in medical oncology. Toxicity was checked at proper intervals following CTCAE V3.0 (Common Terminology Criteria for Adverse Events) grading while efficacy was assessed following RECIST1.0 (response evaluation criteria in solid tumors). The patients were categorized into two groups—i.e., non-responding subjects (reflecting progressive disease) and subjects with clinical benefits (stable disease, showing partial or complete response).
2.7. Pharmacokinetic Sampling
Pharmacokinetic sampling was carried out after almost 2 months of treatment with trastuzumab or 5 courses were administrated to attain steady state. Two samples of blood samples were obtained straightaway on venous blood collection tubes after administering trastuzumab (Cmin, or trough levels) and instantly after infusion (Cmax). These samples of blood were then immediately centrifuged (3000× g, 15 min 4 °C), fractions of plasma were then obtained and maintained at a temperature of −80 °C until further processing.
3. Results
3.1. Method Development
3.1.1. Analytical Procedure for Optimization
During analytical procedure, optimization of both mass spectrometry and chromatographic conditions were carefully optimized to achieve highest possible responses of signature peptides with optimal sensitivity and accuracy. The mass spectrometry (MS)/MS parameters in multiple reaction monitoring (MRM) mode were optimized by injecting 5 μL of the trypsin-digested sample of 2 mg/mL of working standard. The ion-source parameters (source and desolvation temperatures, gas flow rates, and capillary and cone voltage collision energy) were optimized individually to achieve maximum responses. The ion source temperature of 150 °C, desolvation temperature 450 °C, cappilary volatage 3 kV, and cone voltage 40 V showed the best product ion formation with the highest sensitivity during optimization. As far as selecting of column is concern, Acquity UPLC HSS T3 (1.8 μm, 2.1 × 50 mm), Acquity HSS C18 (1.8 μm, 2.1 × 50 mm), and Acquity UPLC BEH Shield C18 (1.7 μm, 2.1 × 50 mm) were evaluated, and among them, the last one produced better separation compared with the rest and was selected for peptide separation. After that, the mobile phase compositions were optimized to maximize peak resolution and signal-to-noise (S/N) ratio. Initially, acetonitrile and water both containing 0.1% formic acid were tried in isocratic and gradient modes of elution. The gradient mode produced better separation for both target and internal standard (IS) peptides which was further optimized to achieve optimum gradient programming with flow rate.
3.1.2. Selection of Signature Peptides
The selection of a unique surrogate peptides is most important for the quantification of proteins by LC-MS/MS analysis. Due to high similarity of amino acid sequences of Mabs, unique and appropriate peptides can only be found in complementarity-determining regions (CDRs) of trastuzumab. During MRM transition optimization, various precursors to product ion of peptides were acquired and screened using MultiQuant software for comparing the peak area responses of various tryptic peptide transitions. Meticulous manual data analysis was performed to filter the peptides and it was observed that fragment crystallizable (Fc) region peptides showed a high degree of sequence similarity to endogenous IgG in the human plasma. Peptide sequences with charge states +2; FTISADTSK (504.772/667.341) and IYPTNGYTR (542.775/808.395) were selected as quantifier peptides for trastuzumab as it produces the highest MS/SM responses. Signature peptides were further confirmed to be located in the fragment antigen-binding (Fab) region by trypsin digestion procedure which uniquely represents the CDR regions of trastuzumab. The spectra representations of signature peptides are shown in Figure 2.
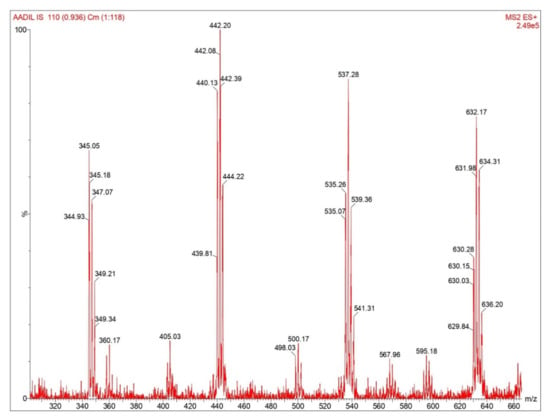
Figure 2.
Spectral representation of signature peptides for trastuzumab quantification.
3.1.3. Digestion Workflow Optimization
The concentration/amount of tris buffer, trypsin, incubation time, and temperature was optimized systematically to achieve recovery of peptides and reproducible and time-effective sample processing. The addition of an alkylation step contributed to an increased and consistent recovery of tryptic peptides. Furthermore, using a SUPER-SELECT HLB SPE cartridge provide µ-elution steps for high-throughput sample preparation. The SPE procedure step (conditioning, washing, and elution) were optimized to achieve consistent recovery and lower matrix effects.
3.2. Method Validation
3.2.1. Selectivity and Matrix Effect
Assay selectivity was determined by screening the blank human plasma samples selected from six different lots. These blank plasma samples were co-processed with the plasma samples spiked at LLOQ level (5 µg/mL) with ISTD. During analysis by UPLC-MS/MS method, no significant interfering peaks in blank samples (compared to LLOQ samples) were observed at the retention time of trastuzumab and ISTD. The representative TIC chromatograms of trastuzumab at LQC level are shown in Figure 3. These findings confirmed that the proposed assay is selective and specific for the quantitative analysis of trastuzumab in human plasma samples and is in compliance with international regulatory guidelines.
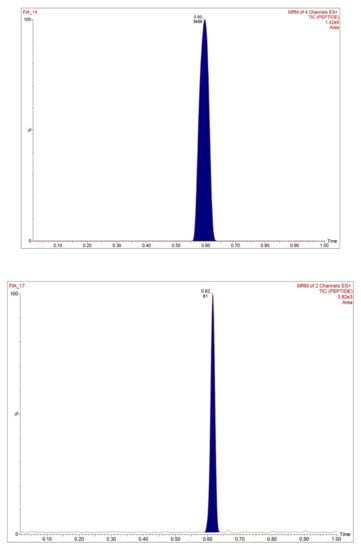
Figure 3.
MRM chromatogram of signature peptides, SILuMAB, illustrating signal in spiked plasma samples.
The matrix effect which was expressed as %ME, was screened in six different lots of human plasma spiked with trastuzumab and ISTD at LQC and HQC levels. The four different aliquots of these samples were spiked in blank post-processed plasma samples and, after analysis, its responses were compared with responses of aqueous samples. Slight ion suppression effects (<15%) have been observed in processed plasma samples with slight variations between them (≤5%). These results indicate that the obvious matrix effects are negligible for analysis of trastuzumab and ISTD.
3.2.2. Linearity and the LLOQ of the Assay
The assay linearity was determined by preparing three different calibration curves (CCs) in blank human plasma spiked with all eight different calibrators (5–500 µg/mL). The CCs were plotted by using the area responses of analyte/ISTD versus nominal concentration of analyte. All CCs were linear with best fitting having a mean correlation coefficient (r2) at 0.9965 (±0.0015) over the concentration ranges. The weighting factor was optimized and 1/x2 showed best results during back calculation of the concentration of the calibrators. The back calculated concentrations of different calibrators of CCs were within the acceptable limit of the ranges. The lowest concentration of the CCs (5 µg/mL) was considered as LLOQ of the assay which was determined with acceptable accuracy and precision and its responses were five times higher than the bank samples.
3.2.3. Precision and Accuracy
The precision (expressed as CV, %) and accuracy (expressed as % difference from nominal values) of trastuzumab in human plasma samples were evaluated at LLOQ and all three QC concentrations (LQC, MQC, and HQC) and are presented in Table 1. The intra-run (within day) and between run (inter-day) were evaluated by using six replications of all four types of samples. The intra-run and inter-run precision of trastuzumab were found to be ≤10.1% ≤ 8.9%, respectively. These results confirmed that the developed assay is accurate and precise and therefore reliable for quantification trastuzumab in plasma samples.

Table 1.
Patients’ characteristics and stage of disease.
3.2.4. Stability
Trastuzumab (mAbs) is a biological group of molecules and therefore its stability under different storage and exposure conditions needs to be assured. All stabilities studied were determined in six replicates using LQC and HQC concentration against freshly prepared CCs. Its stability during freeze–thaw cycles was determined by processing the QC samples after doing three freeze(at −65 ± 10 °C)–thaw (+4 °C) exposures and analyzing the samples (LQC and HQC) against CCs. The short-term stability was determined by processing the samples (LQC and HQC) and keeping them for 8 h for at +4 °C. The auto sampler stability was performed by storing the tryptic digest mixture in the auto-sampler for 96 h, maintaining a temperature of +4 °C. The long-term stability was determined after performing the digestion steps of the samples by keeping them for two months at −65 ± 10 °C. All samples were found to be stable during the analysis of the QC samples after the proposed exposure conditions (Table 2). These results confirmed that the peptides are stable and are less susceptible for degradation at anticipated temperature/storage conditions.

Table 2.
Precision and accuracy values at LLOQ, LQC, MQC, and HQC concentration of trastuzumab in human plasma samples during the course of validation (n = 6).
3.2.5. Robustness
All the parameters needed to check the robustness were checked to improve the method sensitivity and to alleviate the overall robustness. Mobile phase pH, concentrations of additives in the eluent, organic solvent content in the eluent, column temperature, flow rate, drying gas flow, nebulizer gas flow, and ion source configuration were used to evaluate the robustness of the method.
3.3. Toxicity and Efficacy
Following CTCAE V3.0. Criteria, 30% of patients (i.e., 6 out of 20) had reported grade-3 (severe) toxicities which include anemia, thrombocytopenia, mucositis, and nephrotoxicity. Moreover, two patients reported severe radiation induced dermatitis; however, no grade-4 toxicities were reported here. In RECIST 1.0. evaluation, 50% (i.e., 10 out of 20 patients) had progressive disease while the remaining half (50%) were categorized as responding with treatment (i.e., patients with clinical benefit) including 35% patients (7 out of 10) with partial response and 15% patients (3 out of 10 patients) with complete response. Almost half of the patients (9 out of 20 patients) died during the follow-up period. The average survival was almost one year from the start of trastuzumab therapy. Standard t-test was used to calculate the differences between clinical benefit and progressive disease and p-value < 0.05 was considered as statistically significant. In addition, t-test was also used to determine the differences in exposure levels between patients with and patients without severe toxicities. Comparisons of proportions were performed using Fisher’s exact test considering the small number of patients.
3.4. TDM and Exposure Pharmacodynamics Relationships
Out of 20 patients; we are only able to receive five patients’ plasma samples for determination of Cmin and Cmax values. As presented in Table 3 and Table 4, the range of Cmin was 50.3 to 100.3 with mean concentration value of 68.76 µg/mL however the range for Cmax values was 118.2–160.2 µg/mL with mean concentration value of 138.4 µg/mL. Finally, receiver operating characteristic (ROC) curve analysis showed that 52.25 µg/mL concentration was the Cmin threshold predictive of response with an acceptable sensitivity (88.58%) and specificity (79.25%).

Table 3.
Stability of trastuzumab under different temperature and storage conditions.

Table 4.
Residual and maximal concentration (µg/mL) of trastuzumab in breast cancer patients.
4. Discussion
In around 20 percent of breast cancer, HER2 enhancement can be detected and it is linked to invasive phenotype conditions. In metastatic and adjuvant settings, trastuzumab—a mAb HER2 antibody—has a significant clinical advantage. However, after adjuvant therapy with trastuzumab, some patients experience recurrence of the disease and most patients with metastasis do not gain therapeutic benefits or develop refractory disease within 1 year of treatment [18]. A growing hypothesis is that upstream pharmacokinetic problems, particularly low plasma concentration levels, may be the cause of poor responses in some patients treated with normal but inadequate dosing, in addition to possible downstream tumor deregulation [19]. Nothing about the pharmacokinetics of mAbs is known to date [20]. For years, the invention of precise bioanalytical methods for the measurement of plasma concentrations in routine patients has remained a challenging task [21]. Ligand-binding assay, in which most of the methods widely used are related—particularly enzyme-linked immunosorbent assays (ELISA) such as canonical ELISA methods, either as part of clinical registration or in academic research—have long been the standard method for TDM of therapeutic antibodies [22,23]. In addition, some commercial kits are now also available, but only a few clinical pharmacokinetic laboratories in oncology settings or university hospitals have experience with the proper handling of in-house ELISA procedures, which may limit the widespread use of this technique as part of routine bedside monitoring of therapeutic drugs. Moreover, these methods lack the selectivity to distinguish between significant molecular differences (such as point mutations in the target protein or post-translational modifications) and display limited multiplexing capabilities. In this regard, we have established a simple and original UPLC-MS plasma trastuzumab test method that meets the conflicting routine TDM sensitivity, simplicity, solidity, and low-cost requirements. A fast sample preparation workflow with SPE μ-elution technique and sensitive with specificmass spectrometric detection are the basis of this original technique. Efficient quantification of residual concentrations was performed in all test samples with the combination of easy and rapid (i.e., 2 h) pellet digestion and high-pH reverse pellet fractionation prior to low-pH separation LC. The degree of ionization suppression, estimated at only about ≤15%, demonstrates the effectiveness of the fractionation method. It may be linked to the complementarity between high-pH fractionation and low-pH reverse-phase chromatography and optimized wash and elution conditions in the SPE. Quantification based on trastuzumab calibration parameters, processed in a similar manner and at the same time as clinical samples—coupled with SIL-peptide internal standardization at the early stage of the procedure—demonstrated high accuracy and precision, in line with the most recent bioanalytical recommendations, and full analytical range and sensitivity of the method. Moreover, reproducibility, accuracy, and transition by stable isotope dilution protein measurement laboratories and TQD instruments have already been well demonstrated [24,25]. Together with the high accuracy and precision of our assay, it makes the way for easy cross-validation and reproducibility of trastuzumab assays between clinical pharmacokinetic laboratories. This initial approach showed its robustness and full ability to differentiate non-responsive patients from patients with clinical benefit when assessed as part of a pilot study in trastuzumab-treated patients. Based on residual and maximum concentration control, neither population-based identification of individual PK parameters nor absolute exposure level simulations were found, e.g., this technique was used in the actual routine atmosphere of the institute to measure its performance under natural environment conditions. For example, routine care staff conducted sampling, and samples were then taken to the laboratory as routinely used tubes, without complete control of the shelf and the conditions of transport from sample collection to laboratory. However, our strict analytical conditions have never impaired the matrix effect, considering that samples could have remained unattended before being shipped to the laboratory and that regular patients are even more polymedicated than is commonly the case in real clinical trials. Indeed, no inclusion criteria other than trastuzumab therapy have been maintained and a number of stages, co-medications, and sufficient breast cancer localization have been considered. This variety in the pilot study was also not a confounding factor. For example, it has been found that patients with metastatic diseases are equally spread to non-responsive patients and to clinically helpful patients (data not shown). Concomitant radiotherapy or the treatment of carboplatin versus cisplatin, which was linked with 5-FU, which also did not impact clinical outcomes (data not shown). We found that there were extremely variable levels of trastuzumab sensitivity among patients. For example, the mean value of trough level was 68.75 μg/mL with % CV of 30.75%, while mean Cmax level was 138.4 μg/mL with % CV of 14.7%, indicating that variation in residual range concentration of trastuzumab achieved more than the than the median concentrations for other patients at the end of the infusion. Conversely, the median concentration was smaller for some other patients than the residual concentrations found elsewhere. Furthermore, as has already been shown in other populations, the effectiveness was associated with the pharmacokinetics of trastuzumab, as both Cmin—and, to some extent—Cmax levels were higher in stable/responsive patients than in progressive disease patients. A marked difference in Cmin values was first shown and the ROC analysis found that residual concentrations above 52.25 μg/mL, with 88.58% sensitivity and 79.25% specificity, were closely correlated with clinical gain. We have noticed that there was a smaller statistical gap in their respective Cmax values between stable/responsive patients and non-response patients. Considering that the Cmin could be more important than Cmax therapeutic efficacy values for trastuzumab is consistent with the PK/PD relationship of the majority of mAbs used in the treatment of breast tumors. Huge antibodies are difficult to infiltrate solid tumors and it is also widely agreed that it is easy to meet only extracellular targets of the outer cell layer of the tumor mass over time. In laboratory models, it has already been shown, in particular with trastuzumab, that its distribution decreases with the distance from the blood vessels and that there is no antibody in hypoxic regions such as the tumor core [26]. As a result, retaining continuous inhibition of usable tumor surface EGFR targets with prolonged trough levels may be a safer way to optimize effectiveness than to acquire elevated Cmax values and thus significant volumes of plasma trastuzumab that will essentially be unable to penetrate deep tumor tissues. Here, both patients had KRas and NRas wild-type status and could thus respond to anti-EGFR therapy in principle. However, 50% of patients have progressive ailments, given the favorable downstream pharmacogenomic profile. Accordingly, our findings suggest that patients with residual concentrations of trastuzumab below 52 μg/mL are likely to have their dose changed to higher doses to raise dosage levels and increase efficacy, noting that a PK/PD interaction of this type has not been illustrated by toxic cases. However, in this pilot trial, despite efforts to change the dosage, much of the adverse effects we found (i.e., cardiotoxicity) could not be due to anti-EGFR treatment, but rather to related chemotherapy—i.e., cisplatin, carboplatin, and 5-FU [26,27,28,29]. Finally, in this pilot study, the number of patients and the study length were also too limited to determine adequate survival. We found, however, that the mean survival of nine dead patients in the low Cmin level subgroup (four patients) was 6 months while the average survival of those in the high Cmin subgroup (five patients) was 13 months. This finding was consistent with the positive effect we have observed between higher residual trastuzumab concentrations and clinical effectiveness, albeit viewed only as a pattern with no statistical significance. The current dose strategy for oncology and especially targeted therapy, such as mAbs in chemotherapy, is perceived to be sub-optimal for some patients and thus needs better dosing methods for optimal drug dosage for each specific patient. Therefore, individualized TDM-guided dosing with mAb has the ability to be used in clinical oncology [30].
5. Conclusions
In this study, a UPLC-MS/MS assay has been developed and validated for the quantitative determination of trastuzumab in human plasma. The method was successfully applied in the measurement of residual and maximal concentration of trastuzumab in breast cancer patients. While monocentric and performed in a small number of patients, this prospective evidence-of-concept research strongly indicates that monitoring of residual trastuzumab concentrations using the UPLC-MS/MS technique is feasible and inexpensive while providing conclusive evidence to predict clinical efficacy, in addition to standard downstream pharmacogenomics tests at a tumor level. This approach could also be utilized as a convenient approach for implementing a TDM strategy with trastuzumab for early decision-making (i.e., titrating the trastuzumab dose for personalized therapy). Instead of dosing, the TDM could be used to customize the schedule; that is, to determine when the patient will reach the trough concentrations that will lead to efficacy. Eventually, this could reduce drug costs by preventing excessive treatment. TDM with PK modeling and simulation along with randomized prospective trials are needed to verify if precision medicine can be part of immunotherapy [31]. Creating a clinical application of TDM first requires an expansion of the evidence base, then simplifying sampling strategies, such as using dried blood spots or body fluids other than blood. Pharmacometrics that combine pharmacokinetics, individual patient characteristics, and drug concentrations should utilize Bayesian estimations and provide clinics with clinical decision support [32].
Author Contributions
Conceptualization, supervision—O.A. and K.M.; Methodology—A.A.S., R.A.R., M.I. (Mohd Imran) and A.J.A.; Validation—M.I. (Muzaffar Iqbal), S.A. and F.S.; Data curation—A.J.A. and M.M.G.; Funding acquisition—S.A. and M.M.G.; Project administration—O.A. and K.M.; Software—F.S.; Writing—original draft—A.A.S.; Writing—review and editing—M.I. (Muzaffar Iqbal), F.S. and S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The approval from local ethics committee (Scientific Research and Ethics Review Committee Batra Hospital and Research Centre, Tughlaqabad Institutional Area M.B. Road, New Delhi-110062) with letter no. SRERC/2018/19/09 and submission no. SRERC/2018/TH/24 was obtained before the commencement of the study.
Informed Consent Statement
All the patients were informed about the study and informed consent was signed prior to sampling.
Data Availability Statement
This study did not report any data.
Acknowledgments
The authors extend their sincere appreciation to Jamia Hamdard for providing research facilities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Boekhout, A.H.; Beijnen, J.H.; Schellens, J.H. Trastuzumab. Oncologist 2011, 16, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhabi, N.A.; Park, C.G.; Kim, Y.O. LC-MS/MS validation analysis of trastuzumab using dSIL approach for evaluating pharmacokinetics. Molecules 2016, 21, E1464. [Google Scholar]
- Albanell, J.; Codony, J.; Rovira, A.; Mellado, B.; Gascón, P. Mechanism of action of anti-HER2 monoclonal antibodies: Scientific update on trastuzumab and 2C4. In New Trends in Cancer for the 21st Century; Springer: Boston, MA, USA, 2003; pp. 253–268. [Google Scholar]
- Claret, F.X.; Vu, T.T. Trastuzumab: Updated mechanisms of action and resistance in breast cancer. Front. Oncol. 2012, 18, E62. [Google Scholar]
- Siegel, J.P. Biologics license application for trastuzumab. Clin. Rev. 1998, 98, 3–49. [Google Scholar]
- OGIVRI (Trastuzumab) Health Canada Product Information. Available online: https://health-products.canada.ca/dpd-bdpp/info.do?lang=en&code=96453 (accessed on 12 September 2020).
- KANJINTI (Trastuzumab-Anns) Prescribing Information USFDA Document. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/761073s000lbl.pdf (accessed on 12 September 2020).
- Fusaro, V.A.; Mani, D.R.; Mesirov, J.P.; Carr, S.A. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat. Biotechnol. 2009, 27, 190–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oude Munnink, T.H.; Henstra, M.J.; Segerink, L.I.; Movig, K.L.; Brummelhuis-Visser, P. Therapeutic drug monitoring of monoclonal antibodies in inflammatory and malignant disease: Translating TNF-α experience to oncology. Clin. Pharmacol. Ther. 2016, 99, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Ladwig, P.M.; Barnidge, D.R.; Willrich, M.A. Mass spectrometry approaches for identification and quantitation of therapeutic monoclonal antibodies in the clinical laboratory. Clin. Vaccine Immunol. 2017, 24, E00545-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willeman, T.; Jourdil, J.F.; Gautier-Veyret, E.; Bonaz, B.; Stanke-Labesque, F. A multiplex liquid chromatography tandem mass spectrometry method for the quantification of seven therapeutic monoclonal antibodies: Application for adalimumab therapeutic drug monitoring in patients with Crohn’s disease. Anal. Chim. Acta 2019, 1067, 63–70. [Google Scholar] [CrossRef]
- Damen, C.W.; de Groot, E.R.; Heij, M.; Boss, D.S.; Schellens, J.H.; Rosing, H.; Beijnen, J.H.; Aarden, L.A. Development and validation of an enzyme-linked immunosorbent assay for the quantification of trastuzumab in human serum and plasma. Anal. Biochem. 2009, 391, 114–120. [Google Scholar] [CrossRef]
- Sanchez, A.B.; Nguyen, T.; Dema-Ala, R.; Kummel, A.C.; Kipps, T.J.; Messmer, B.T. A general process for the development of peptide-based immunoassays for monoclonal antibodies. Cancer Chemother. Pharmacol. 2010, 66, 919–925. [Google Scholar] [CrossRef] [Green Version]
- Becher, F.; Ciccolini, J.; Imbs, D.C.; Marin, C.; Fournel, C.; Dupuis, C.; Fakhry, N.; Pourroy, B.; Ghettas, A.; Pruvost, A.; et al. A simple and rapid LC-MS/MS method for therapeutic drug monitoring of cetuximab: A GPCO-UNICANCER proof of concept study in head-and-neck cancer patients. Sci. Rep. 2017, 7, E2714. [Google Scholar] [CrossRef] [PubMed]
- El Amrani, M.; Donners, A.A.; Hack, C.E.; Huitema, A.D.; van Maarseveen, E.M. Six-step workflow for the quantification of therapeutic monoclonal antibodies in biological matrices with liquid chromatography mass spectrometry—A tutorial. Anal. Chim. Acta 2019, 10180, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration, Center for Drug Evaluation and Research; Center for Veterinary Medicine (CVM). Bioanalytical Method Validation Guidence for Industry. 2018. Available online: https://www.fda.gov/files/drugs/published/Bioanalytical-Method-Validation-Guidance-for-Industry.pdf (accessed on 18 October 2020).
- Duggan, J.X.; Vazvaei, F.; Jenkins, R. Bioanalytical method validation considerations for LC-MS/MS assays of therapeutic proteins. Bioanalysis 2015, 7, 1389–1395. [Google Scholar] [CrossRef]
- Gajria, D.; Chandarlapaty, S. HER2-amplified breast cancer: Mechanisms of trastuzumab resistance and novel targeted therapies. Expert Rev. Anticancer Ther. 2011, 11, 263–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- André, F.; Ciccolini, J.; Spano, J.P.; Penault-Llorca, F.; Mounier, N.; Freyer, G.; Blay, J.Y.; Milano, G. Personalized medicine in oncology: Where have we come from and where are we going? Pharmacogenomics 2013, 14, 931–939. [Google Scholar] [CrossRef]
- Keizer, R.J.; Huitema, A.D.; Schellens, J.H.; Beijnen, J.H. Clinical pharmacokinetics of therapeutic monoclonal antibodies. Clin. Pharmacokinet. 2010, 49, 493–507. [Google Scholar] [CrossRef] [PubMed]
- Damen, C.W.; Schellens, J.H.; Beijnen, J.H. Bioanalytical methods for the quantification of therapeutic monoclonal antibodies and their application in clinical pharmacokinetic studies. Hum. Antib. 2009, 18, 47–73. [Google Scholar] [CrossRef]
- Suárez, I.; Salmerón-García, A.; Cabeza, J.; Capitán-Vallvey, L.F.; Navas, N. Development and use of specific ELISA methods for quantifying the biological activity of bevacizumab, cetuximab and trastuzumab in stability studies. J. Chromatogr. B. 2016, 1032, 155–164. [Google Scholar] [CrossRef]
- Spengler, M.; Adler, M.; Niemeyer, C.M. Highly sensitive ligand-binding assays in pre-clinical and clinical applications: Immuno-PCR and other emerging techniques. Analyst 2015, 140, 6175–6194. [Google Scholar] [CrossRef] [Green Version]
- Abbatiello, S.E.; Schilling, B.; Mani, D.R.; Zimmerman, L.J.; Hall, S.C.; MacLean, B.; Albertolle, M.; Allen, S.; Burgess, M.; Cusack, M.P.; et al. Large-scale interlaboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer-relevant proteins in plasma. Mol. Cell Proteom. 2015, 14, 2357–2374. [Google Scholar] [CrossRef] [Green Version]
- Addona, T.A.; Abbatiello, S.E.; Schilling, B.; Skates, S.J.; Mani, D.R.; Bunk, D.M.; Spiegelman, C.H.; Zimmerman, L.J.; Ham, A.-J.L.; Keshishian, H.; et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 2009, 27, 633–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.M.; Tannock, I.F. The distribution of the therapeutic monoclonal antibodies cetuximab and trastuzumab within solid tumors. BMC Cancer 2010, 10, E255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passot, C.; Azzopardi, N.; Renault, S.; Baroukh, N.; Arnoult, C.; Ohresser, M.; Boisdron-Celle, M.; Gamelin, E.; Watier, H.; Paintaud, G.; et al. Influence of FCGRT gene polymorphisms on pharmacokinetics of therapeutic antibodies. MAbs 2013, 5, 614–619. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.G.; Ciccolini, J.; Blesius, A.; Dahan, L.; Bagarry-Liegey, D.; Brunet, C.; Varoquaux, A.; Frances, N.; Marouani, H.; Giovanni, A.; et al. DPD-based adaptive dosing of 5-FU in patients with head and neck cancer: Impact on treatment efficacy and toxicity. Cancer Chemother. Pharmacol. 2011, 67, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Minasian, L.; Rosen, O.; Auclair, D.; Rahman, A.; Pazdur, R.; Schilsky, R.L. Optimizing dosing of oncology drugs. Clin. Pharmacol. Ther. 2014, 96, 572–579. [Google Scholar] [CrossRef]
- Gao, B.; Yeap, S.; Clements, A.; Balakrishnar, B.; Wong, M.; Gurney, H. Evidence for therapeutic drug monitoring of targeted anticancer therapies. J. Clin. Oncol. 2012, 30, 4017–4025. [Google Scholar] [CrossRef]
- Chatelut, E.; Hendrikx, J.J.; Martin, J.; Ciccolini, J.; Moes, D.J. Unraveling the complexity of therapeutic drug monitoring for monoclonal antibody therapies to individualize dose in oncology. Pharmacol. Res. Persp. 2021, 9, E00757. [Google Scholar] [CrossRef]
- Martin, J.H.; Olver, I. Precision medicine-based drug treatment individualization in oncology. Br. J. Clin. Pharmacol. 2021, 87, 223–226. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).