Construction of Recombinant Magnetospirillum Strains for Nitrate Removal from Wastewater Based on Magnetic Adsorption
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Cell Growth and Magnetic Response
2.3. Construction of mgfnr Mutant Strain Δmgfnr
2.4. Transmission Electron Microscopic (TEM) Observation
2.5. Quantitative Real-Time Reverse Transcription PCR (qRT-PCR)
2.6. Electrophoretic Mobility Shift Assays (EMSAs)
2.7. Iron Absorption Capability
2.8. Protein Expression Vector Construction and Anaerobic Purification
2.9. Chromatin Immunoprecipitation-Quantitative PCR (ChIP-qPCR)
2.10. Total Nitrogen Content
3. Results
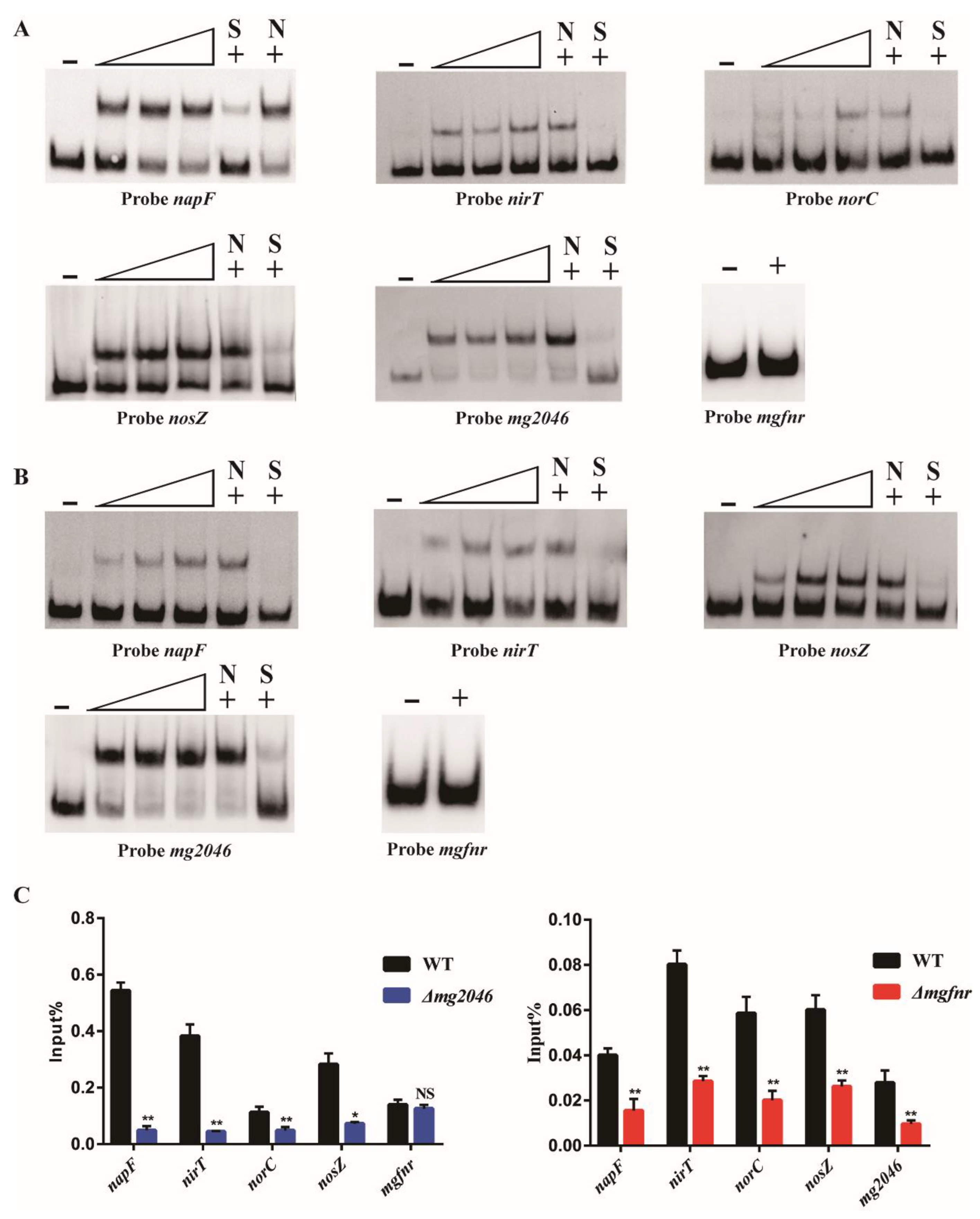
3.1. Combined Effects of Dissimilatory Denitrification Pathway Genes Determined by EMSAs and ChIP-qPCR
3.2. Transcription Levels of Dissimilatory Denitrification Pathway Genes
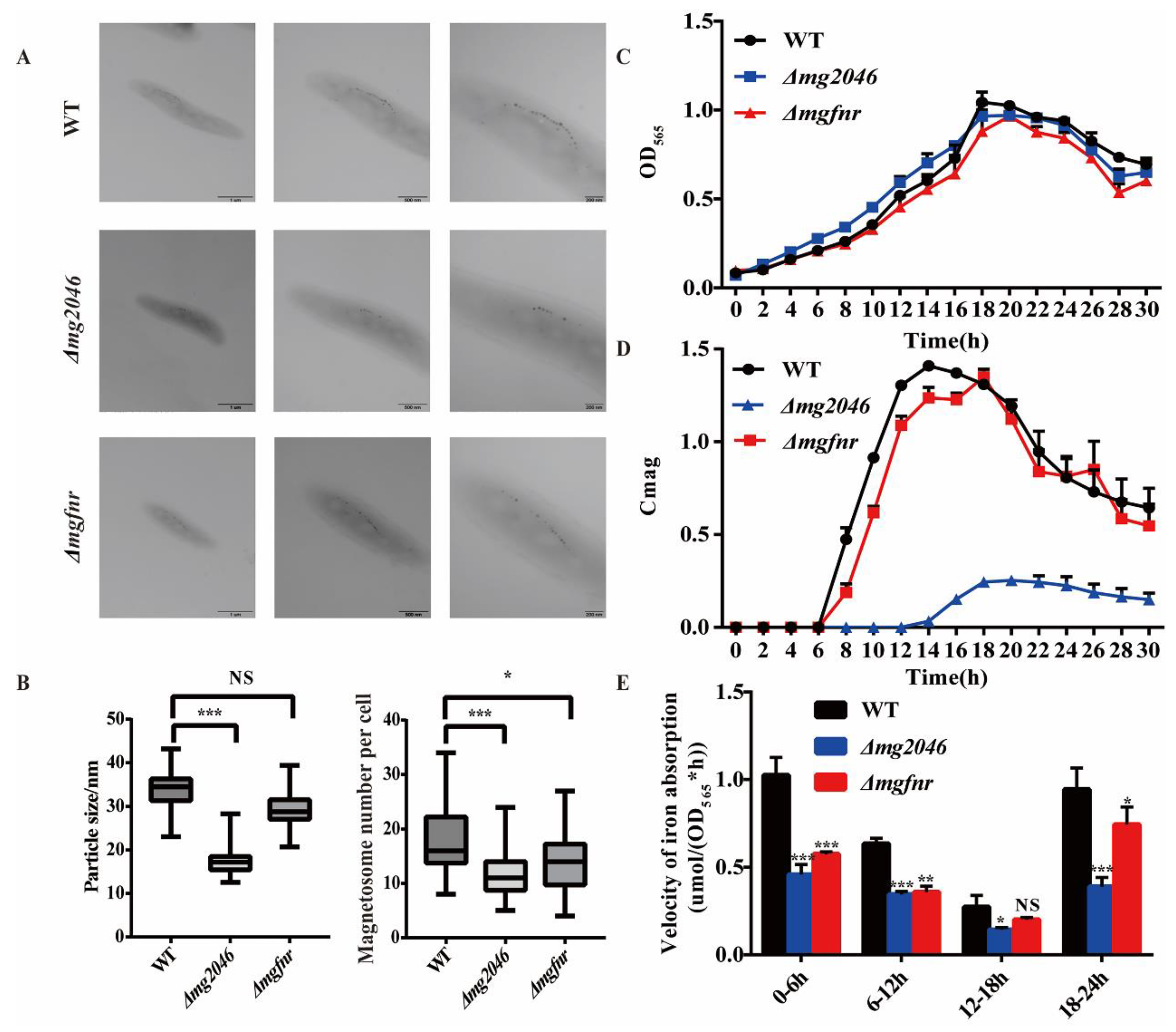
3.3. Phenotypic Analysis of WT and Δmg2046/Δmgfnr
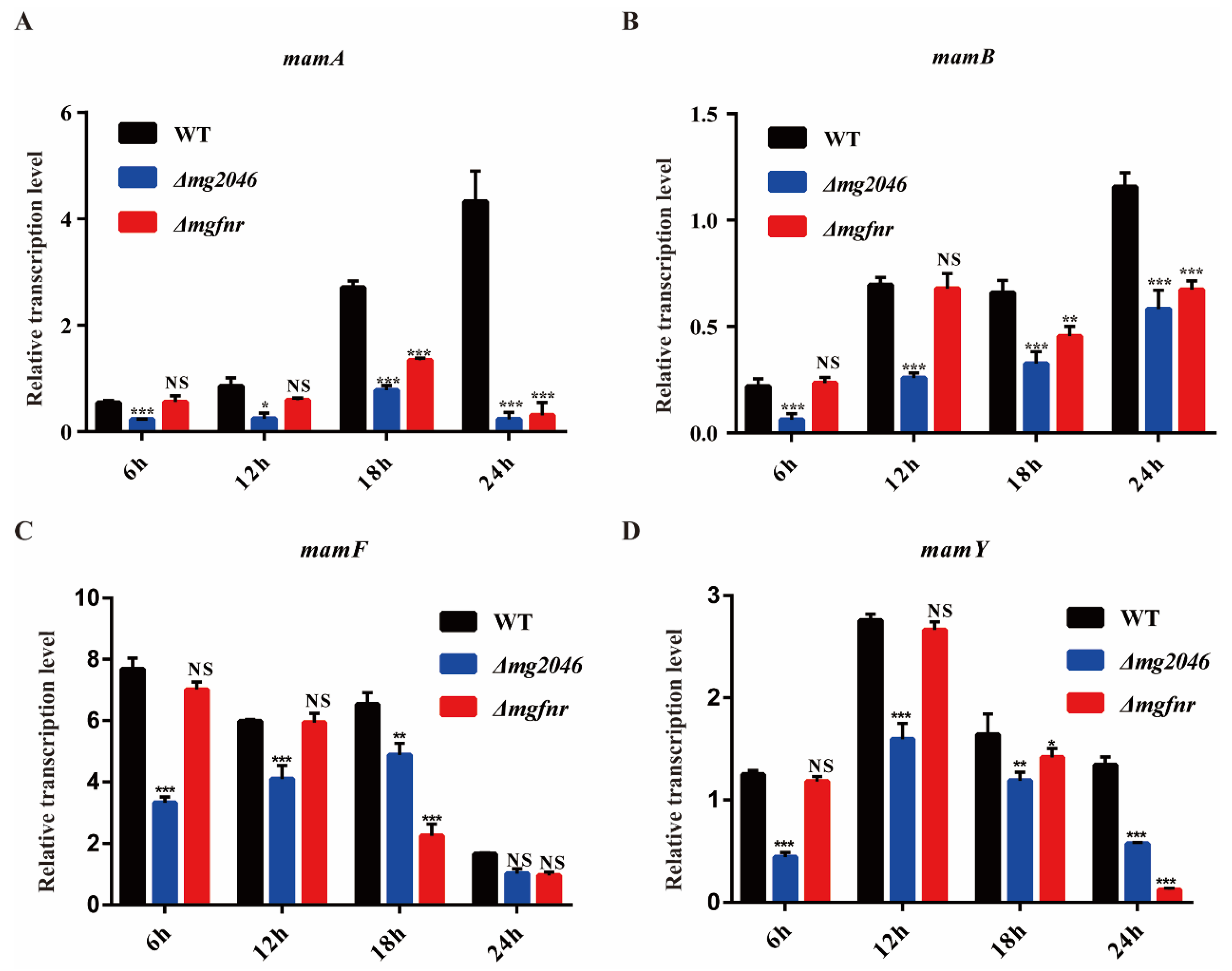
3.4. Transcription of Genes Involved in Magnetosome Synthesis
3.5. Comparative Utilization of Various Nitrogen Sources by WT and Δmg2046/Δmgfnr
3.6. Comparative Utilization of Nitrogen Sources from Synthetic Wastewater by WT and Δmg2046/Δmgfnr
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wick, K.; Heumesser, C.; Schmid, E. Groundwater nitrate contamination: Factors and indicators. J. Environ. Manag. 2012, 111, 178–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fields, S. Global nitrogen-Cycling out of control. Environ. Health Perspect. 2004, 112, A556–A563. [Google Scholar] [CrossRef] [PubMed]
- Trodden, W.; O’Boyle, S. Water Quality in 2020: An Indicators Reports; EPA: Wexford, Ireland, 2020.
- Park, J.Y.; Yoo, Y.J. Biological nitrate removal in industrial wastewater treatment: Which electron donor we can choose. Appl. Microbiol. Biotechnol. 2009, 82, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Li, X.M.; Fan, W.H.; Wang, J.L. Heterotrophic nitrification and aerobic denitrification by a novel Acinetobacter sp. ND7 isolated from municipal activated sludge. Bioresour. Technol. 2020, 301, 8. [Google Scholar] [CrossRef]
- Elefsiniotis, P.; Wareham, D.G.; Smith, M.O. Use of volatile fatty acids from an acid-phase digester for denitrification. J. Biotechnol. 2004, 114, 289–297. [Google Scholar] [CrossRef]
- Sahinkaya, E.; Kilic, A.; Duygulu, B. Pilot and full scale applications of sulfur-based autotrophic denitrification process for nitrate removal from activated sludge process effluent. Water Res. 2014, 60, 210–217. [Google Scholar] [CrossRef]
- Tang, Y.; Ziv-El, M.; Zhou, C.; Shin, J.H.; Ahn, C.H.; Meyer, K.; Candelaria, D.; Friese, D.; Overstreet, R.; Scott, R.; et al. Bioreduction of nitrate in groundwater using a pilot-scale hydrogen-based membrane biofilm reactor. Front. Environ. Sci. Eng. China 2010, 4, 280–285. [Google Scholar] [CrossRef]
- Wang, J.L.; Jing, K. The characteristics of anaerobic ammonium oxidation (ANAMMOX) by granular sludge from an EGSB reactor. Process Biochem. 2005, 40, 1973–1978. [Google Scholar]
- Wang, J.L.; Yang, N. Partial nitrification under limited dissolved oxygen conditions. Process Biochem. 2004, 39, 1223–1229. [Google Scholar]
- Wang, J.L.; Chu, L.B. Biological nitrate removal from water and wastewater by solid-phase denitrification process. Biotechnol. Adv. 2016, 34, 1103–1112. [Google Scholar] [CrossRef]
- Di Capua, F.; Pirozzi, F.; Lens, P.N.L.; Esposito, G. Electron donors for autotrophic denitrification. Chem. Eng. J. 2019, 362, 922–937. [Google Scholar] [CrossRef]
- Hwang, Y.; Mines, P.D.; Jakobsen, M.H.; Andersen, H.R. Simple colorimetric assay for dehalogenation reactivity of nanoscale zero-valent iron using 4-chlorophenol. Appl. Catal. B-Environ. 2015, 166, 18–24. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Wang, X.M.; Yang, H.W.; Xie, Y.F.F. Haloacetic acid removal by sequential zero-valent iron reduction and biologically active carbon degradation. Chemosphere 2013, 90, 1563–1567. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Wang, J. Various electron donors for biological nitrate removal: A review. Sci. Total Environ. 2021, 794, 148699. [Google Scholar] [CrossRef]
- Lefevre, C.T.; Bazylinski, D.A. Ecology, Diversity, and Evolution of Magnetotactic Bacteria. Microbiol. Mol. Biol. Rev. 2013, 77, 497–526. [Google Scholar] [CrossRef] [Green Version]
- Lou, X.; Chen, Z.; He, Z.; Sun, M.; Sun, J. Bacteria-Mediated Synergistic Cancer Therapy: Small Microbiome Has a Big Hope. Nano-Micro Lett. 2021, 13, 1–26. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, Y.Z.; Ding, X.H.; Liu, Y.; Shen, F.; Zhang, X.H.; Deng, S.H.; Xiao, H.; Yang, G.; Peng, H. Magnetotactic bacteria: Promising biosorbents for heavy metals. Appl. Microbiol. Biotechnol. 2012, 95, 1097–1104. [Google Scholar] [CrossRef]
- Li, Y.J.; Katzmann, E.; Borg, S.; Schuler, D. The Periplasmic Nitrate Reductase Nap Is Required for Anaerobic Growth and Involved in Redox Control of Magnetite Biomineralization in Magnetospirillum gryphiswaldense. J. Bacteriol. 2012, 194, 4847–4856. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Nava, Y.; Maranon, E.; Soons, J.; Castrillon, L. Denitrification of high nitrate concentration wastewater using alternative carbon sources. J. Hazard. Mater. 2010, 173, 682–688. [Google Scholar] [CrossRef]
- Rong, C.B.; Huang, Y.J.; Zhang, W.J.; Jiang, W.; Li, Y.; Li, J.L. Ferrous iron transport protein B gene (feoB1) plays an accessory role in magnetosome formation in Magnetospirillum gryphiswaldense strain MSR-1. Res. Microbiol. 2008, 159, 530–536. [Google Scholar] [CrossRef]
- Dailey, H.A.; Lascelles, J. Reduction of iron and synthesis of protoheme by spirillum-itersonii and other organisms. J. Bacteriol. 1977, 129, 815–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wang, Q.; Zhang, Y.; Wang, Y.; Zhou, Y.; Zhang, W.; Wen, T.; Li, L.; Zuo, M.; Zhang, Z.; et al. Transcriptome analysis reveals physiological characteristics required for magnetosome formation in Magnetospirillum gryphiswaldense nMSR-1. Environ. Microbiol. Rep. 2016, 8, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zheng, H.L.; Wang, Q.; Jiang, W.; Wen, Y.; Tian, J.S.; Sun, J.B.; Li, Y.; Li, J.L. Novel Protein Mg2046 Regulates Magnetosome Synthesis in Magnetospirillum gryphiswaldense MSR-1 by Modulating a Proper Redox Status. Front. Microbiol. 2019, 10, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.J.; Bali, S.; Borg, S.; Katzmann, E.; Ferguson, S.J.; Schuler, D. Cytochrome cd(1) Nitrite Reductase NirS Is Involved in Anaerobic Magnetite Biomineralization in Magnetospirillum gryphiswaldense and Requires NirN for Proper d(1) Heme Assembly. J. Bacteriol. 2013, 195, 4297–4309. [Google Scholar] [CrossRef] [Green Version]
- Hao, X.D.; Furumai, H.; Chen, G.H. Resource recovery: Efficient approaches to sustainable water and wastewater treatment. Water Res. 2015, 86, 83–84. [Google Scholar] [CrossRef]
- Cheng, C.; Zhou, Z.; Pang, H.J.; Zheng, Y.; Chen, L.Y.; Jiang, L.M.; Zhao, X.D. Correlation of microbial community structure with pollutants removal, sludge reduction and sludge characteristics in micro-aerobic side-stream reactor coupled membrane bioreactors under different hydraulic retention times. Bioresour. Technol. 2018, 260, 177–185. [Google Scholar] [CrossRef]
- Hussain, A.; Lee, J.; Reid, R.; Lee, H.S. Upflow anaerobic-microaerobic fixed biofilm reactor integrating methanogenesis with partial nitrification. Chem. Eng. J. 2018, 348, 281–291. [Google Scholar] [CrossRef]
- Meng, J.; Li, J.L.; Li, J.Z.; Antwi, P.; Deng, K.W.; Wang, C.; Buelna, G. Nitrogen removal from low COD/TN ratio manure-free piggery wastewater within an upflow microaerobic sludge reactor. Bioresour. Technol. 2015, 198, 884–890. [Google Scholar] [CrossRef]
- Mesquita, D.P.; Amaral, A.L.; Ferreira, E.C. Activated sludge characterization through microscopy: A review on quantitative image analysis and chemometric techniques. Anal. Chim. Acta 2013, 802, 14–28. [Google Scholar] [CrossRef]
- Murat, D.; Quinlan, A.; Vali, H.; Komeili, A. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc. Natl. Acad. Sci. USA 2010, 107, 5593–5598. [Google Scholar] [CrossRef] [Green Version]
- Verlicchi, P.; Al Aukidy, M.; Zambello, E. Occurrence of pharmaceutical compounds in urban wastewater: Removal, mass load and environmental risk after a secondary treatment-A review. Sci. Total Environ. 2012, 429, 123–155. [Google Scholar] [CrossRef] [PubMed]
- Verlicchi, P.; Zambello, E. How efficient are constructed wetlands in removing pharmaceuticals from untreated and treated urban wastewaters? A review. Sci. Total Environ. 2014, 470, 1281–1306. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.J.; Dai, X.H. Sewage sludge-based functional nanomaterials: Development and applications. Environ. Sci.-Nano 2017, 4, 17–26. [Google Scholar] [CrossRef]
- Gonzalez, A.; Hendriks, A.; van Lier, J.B.; de Kreuk, M. Pre-treatments to enhance the biodegradability of waste activated sludge: Elucidating the rate limiting step. Biotechnol. Adv. 2018, 36, 1434–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhen, G.Y.; Lu, X.Q.; Kato, H.; Zhao, Y.C.; Li, Y.Y. Overview of pretreatment strategies for enhancing sewage sludge disintegration and subsequent anaerobic digestion: Current advances, full-scale application and future perspectives. Renew. Sustain. Energy Rev. 2017, 69, 559–577. [Google Scholar] [CrossRef]
- Lu, D.; Wu, D.; Qian, T.T.; Jiang, J.K.; Cao, S.B.; Zhou, Y. Liquid and solids separation for target resource recovery from thermal hydrolyzed sludge. Water Res. 2020, 171, 115476. [Google Scholar] [CrossRef]



| Primer | Sequence (5′-3′) | Description |
|---|---|---|
| Qrpoc-F | ATCTGGTCTACCGCCATTG | qRT-PCR for rpoc gene |
| Qrpoc-R | CCTTGCCGAACGAAATACC | qRT-PCR for rpoc gene |
| QmamA-F | GCCTATCCGTGGCGAAGAA | qRT-PCR for mamA gene |
| QmamA-R | TCGGCATCGTAAACCTGCT | qRT-PCR for mamA gene |
| QmamB-F | AGGTCGTGTGGTGGGCAT | qRT-PCR for mamB gene |
| QmamB-R | CGCTCATCCGCAGGCTTA | qRT-PCR for mamB gene |
| Qmms6-F | GGTTGGCGTTGGGAAGGT | qRT-PCR for mms6 gene |
| Qmms6-R | CATCGCTCTGTGCCGCTT | qRT-PCR for mms6 gene |
| QmmsF-F | TCGGGACGACGAGTTTGTC | qRT-PCR for mmsF gene |
| QmmsF-R | GGAACACCACGGAGACCAA | qRT-PCR for mmsF gene |
| QnapF-F | TGATGTCGCACAGCCTTAG | qRT-PCR for napF gene |
| QnapF-R | TGATGTCGCACAGCCTTAG | qRT-PCR for napF gene |
| QnirT-F | CCATTCACTACACCAACCGTTC | qRT-PCR for nirT gene |
| QnirT-R | ATGGCAGTTGCGGCATTC | qRT-PCR for nirT gene |
| QnorC-F | CGGTGTTCGTTGCCTTGA | qRT-PCR for norC gene |
| QnorC-R | CAGACATTGCCCAGTTCCG | qRT-PCR for norC gene |
| QnosZ-F | TCGCCACGGTGTCCTTT | qRT-PCR for nosZ gene |
| QnosZ-R | ATCACCTGACCGCTTTGGC | qRT-PCR for nosZ gene |
| Qmg2046-F | GCTCCATACCCAATGACGC | qRT-PCR for mg2046 gene |
| Qmg2046-R | TGTCCACATCCTCGCCC | qRT-PCR for mg2046 gene |
| Qmgfnr-F | GAGTTGAACCACGACGAAATCA | qRT-PCR for mgfnr gene |
| Qmgfnr-R | CGAACATCTCGCCCGAAA | qRT-PCR for mgfnr gene |
| mgfnrSF-EcoRI | CGGAATTCACCCTGACCGTGGGCAAGCCGGAA | Amplification upstream of mgfnr |
| mgfnrSR-SacI | CGAGCTCACCTTGTGATCGTCGTAATCC | Amplification upstream of mgfnr |
| mgfnrR-SacI | GCTGTTGTTCTTCCTGCT | Confirmation of mgfnr mutant |
| mgfnrF-SacI | TCCACCGAAATGAAACCG | Confirmation of mgfnr mutant |
| mgfnrXF-SacI | CGAGCTCTACCCAGTTGAAGCGTGAAG | Amplification downstream of mgfnr |
| mgfnrXR-XbaI | GCTCTAGAGAAATCGGAAAACAGCCCCA | Amplification downstream of mgfnr |
| Pmg2046-F | GGAATTC ATGACGACGATGATCCA | Expression of mg2046 protein |
| Pmg2046-R | CCTCGAG TTAAACGTTCTCCCATC | Expression of mg2046 protein |
| Pmgfnr-F | CCGGAATTCGTGATCCCCATGCCGCC | Expression of mgfnr protein |
| Pmgfnr-R | CCGCTCGAGCTAATGCGCCCCGCCGC | Expression of mgfnr protein |
| Erpoc-F | TGAAGGAAGCCAAGGACCT | EMSA for rpoc gene promoter |
| Erpoc-R | CGAGGGACGG GTCAAATCCC | EMSA for rpoc gene promoter |
| EnapF-F | CGGCGGTCAAGAAGATGA | EMSA for nap operon |
| EnapF-R | GAGTGCGCCCGAACAAGG | EMSA for nap operon |
| EnirT-F | AAGCAGCAGGGCGTTCCT | EMSA for nir operon |
| EnirT-R | AGTATTTTCATTTTGGACA | EMSA for nir operon |
| EnorC-F | AACCGATCTCATCGGCGAA | EMSA for nor operon |
| EnorC-R | TACCGCCATAAAAGATATT | EMSA for nor operon |
| EnosZ-F | AGACGTCGGGGCAGAAGGT | EMSA for nos operon |
| EnosZ-R | AGCGCGCCAAAGGACACCGT | EMSA for nos operon |
| Emg2046-F | TGTTGGGCGAAATCCTCGT | EMSA for mg2046 gene promoter |
| Emg2046-R | GGCCAGGATG TCCACATCCT | EMSA for mg2046 gene promoter |
| Emgfnr-F | TGGCTGAAATCTGCGAGGTT | EMSA for mgfnr gene promoter |
| Emgfnr-R | TCACGGGCGA CCTTGTGAT | EMSA for mgfnr gene promoter |
| Strain or Plasmid | Description | Source or Reference |
|---|---|---|
| E. coli DH5α | endA1 hsdR17 [r-m+] supE44 thi-1 recA1 gyrA [NalR] relA relA1 Δ[lacZYA-argF] U169 deoR [Ø80Δ (LacZ) M15] | Novagen |
| E. coli DH5α-mgfnrS | DH5α containing pMD18-T-mgfnrS, Ampr | This study |
| E. coli DH5α-mgfnrX | DH5α containing pMD18-T-mgfnrX, Ampr | This study |
| E. coli S17-1 | Thi endA recA hsdR with RP4-2-Tc::Mu-Km::Tn7 integrated in chromosome; Smr, Tra | Novagen |
| E. coli S17-1-Δmgfnr | S17-1 containing pUX-19-mgfnrS-Gm-mgfnrX, Kmr, Gmr | This study |
| E. coli BL21 (DE3) | F- ompT hdSB (rB− mB−) gal dcm (DE3), general purpose expression host | Novagen |
| E. coli BL21-Pmg2046 | BL21 containing pET28a (+)-Pmg2046, Kmr | This study |
| E. coli BL21-Pmgfnr | BL21 containing pET28a (+)-Pmgfnr, Kmr | This study |
| MSR-1 WT | WT M. gryphiswaldense, Nxr | DSM 6361 |
| MSR-1 Δmg2046 | mg2046-deficient mutant, Nxr, Gmr | This study |
| MSR-1 Δmgfnr | mg2046-deficient mutant, Nxr, Gmr | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, H.; Pang, B.; Li, S.; Ma, S.; Xu, J.; Wen, Y.; Tian, J. Construction of Recombinant Magnetospirillum Strains for Nitrate Removal from Wastewater Based on Magnetic Adsorption. Processes 2022, 10, 591. https://doi.org/10.3390/pr10030591
Zheng H, Pang B, Li S, Ma S, Xu J, Wen Y, Tian J. Construction of Recombinant Magnetospirillum Strains for Nitrate Removal from Wastewater Based on Magnetic Adsorption. Processes. 2022; 10(3):591. https://doi.org/10.3390/pr10030591
Chicago/Turabian StyleZheng, Haolan, Bo Pang, Shuli Li, Shijiao Ma, Junjie Xu, Ying Wen, and Jiesheng Tian. 2022. "Construction of Recombinant Magnetospirillum Strains for Nitrate Removal from Wastewater Based on Magnetic Adsorption" Processes 10, no. 3: 591. https://doi.org/10.3390/pr10030591
APA StyleZheng, H., Pang, B., Li, S., Ma, S., Xu, J., Wen, Y., & Tian, J. (2022). Construction of Recombinant Magnetospirillum Strains for Nitrate Removal from Wastewater Based on Magnetic Adsorption. Processes, 10(3), 591. https://doi.org/10.3390/pr10030591





