Synthetic Biology: A New Era in Hydrocarbon Bioremediation
Abstract
:1. Introduction
1.1. Petroleum Composition: Crude Oil
1.2. Traditional Hydrocarbon Bioremediation Techniques
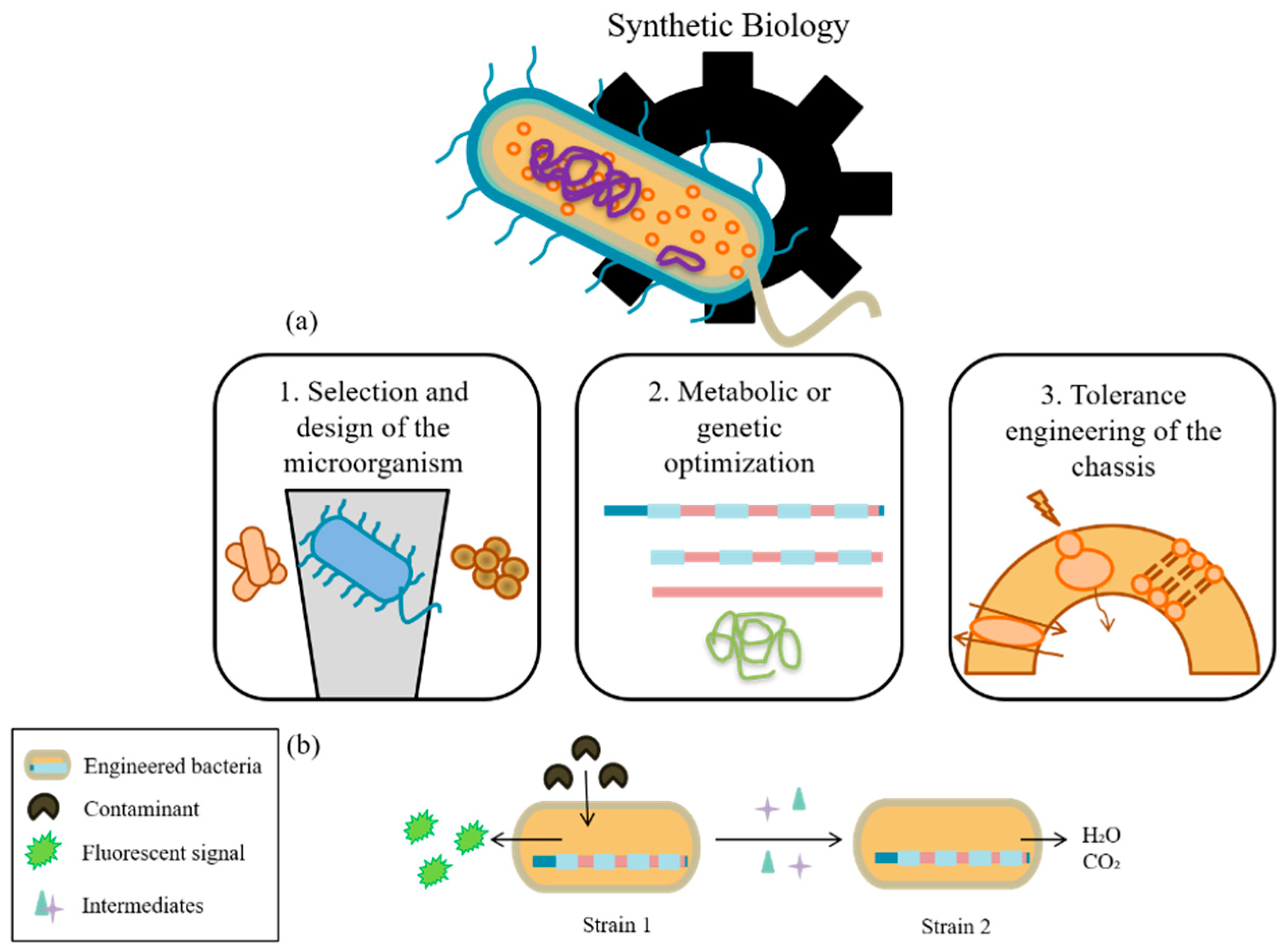
2. Synthetic Biology and Bioremediation
3. Genetically Engineered Microorganisms
4. Biosensors
5. Construction of Synthetic Consortia
6. Risk Assessment of Synthetic Biology
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Speight, J.G.; El-Gendy, N.S. (Eds.) Chapter 1—Petroleum Composition and Properties. In Introduction to Petroleum Biotechnology; Gulf Professional Publishing: Boston, MA, USA, 2018; pp. 1–39. [Google Scholar]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Selley, R.C.; Sonnenberg, S.A. (Eds.) Chapter 2—The Physical and Chemical Properties of Petroleum. In Elements of Petroleum Geology, 3rd ed.; Academic Press: Boston, MA, USA, 2015; pp. 13–39. [Google Scholar]
- Bilal, M.; Iqbal, H.M.N. Microbial-derived biosensors for monitoring environmental contaminants: Recent advances and future outlook. Process Saf. Environ. Prot. 2019, 124, 8–17. [Google Scholar] [CrossRef]
- Wu, M.; Wu, J.; Zhang, X.; Ye, X. Effect of bioaugmentation and biostimulation on hydrocarbon degradation and microbial community composition in petroleum-contaminated loessal soil. Chemosphere 2019, 237, 124456. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Li, G.; Wen, L.; Su, C.; Liu, Y.; Tang, H.; Dai, J. Biodegradation of aromatic pollutants meets synthetic biology. Synth. Syst. Biotechnol. 2021, 6, 153–162. [Google Scholar] [CrossRef]
- Baniasadi, M.; Mousavi, S.M. A Comprehensive Review on the Bioremediation of Oil Spills. In Microbial Action on Hydrocarbons; Kumar, V., Kumar, M., Prasad, R., Eds.; Springer: Singapore, 2018; pp. 223–254. [Google Scholar]
- Jaiswal, S.; Shukla, P. Alternative Strategies for Microbial Remediation of Pollutants via Synthetic Biology. Front. Microbiol. 2020, 11, 808. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Chaudhary, D.R. 21—Engineered microbes and evolving plastic bioremediation technology. In Bioremediation of Pollutants; Pandey, V.C., Singh, V., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 417–443. [Google Scholar]
- Imam, A.; Suman, S.K.; Ghosh, D.; Kanaujia, P.K. Analytical approaches used in monitoring the bioremediation of hydrocarbons in petroleum-contaminated soil and sludge. TrAC Trends Anal. Chem. 2019, 118, 50–64. [Google Scholar] [CrossRef]
- Pal, A.K.; Singh, J.; Soni, R.; Tripathi, P.; Kamle, M.; Tripathi, V.; Kumar, P. 10—The role of microorganism in bioremediation for sustainable environment management. In Bioremediation of Pollutants; Pandey, V.C., Singh, V., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 227–249. [Google Scholar]
- Azubuike, C.C.; Chikere, C.B.; Okpokwasili, G.C. Bioremediation techniques-classification based on site of application: Principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 2016, 32, 180. [Google Scholar] [CrossRef] [Green Version]
- Myazin, V.A.; Korneykova, M.V.; Chaporgina, A.A.; Fokina, N.V.; Vasilyeva, G.K. The Effectiveness of Biostimulation, Bioaugmentation and Sorption-Biological Treatment of Soil Contaminated with Petroleum Products in the Russian Subarctic. Microorganisms 2021, 9, 1722. [Google Scholar] [CrossRef]
- Ossai, I.C.; Ahmed, A.; Hassan, A.; Hamid, F.S. Remediation of soil and water contaminated with petroleum hydrocarbon: A review. Environ. Technol. Innov. 2020, 17, 100526. [Google Scholar] [CrossRef]
- Sakshi; Singh, S.K.; Haritash, A.K. Polycyclic aromatic hydrocarbons: Soil pollution and remediation. Int. J. Environ. Sci. Technol. 2019, 16, 6489–6512. [Google Scholar] [CrossRef]
- Yaman, C. Performance and Kinetics of Bioaugmentation, Biostimulation, and Natural Attenuation Processes for Bioremediation of Crude Oil-Contaminated Soils. Processes 2020, 8, 883. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, R.; Rao, P.; Wu, B.; Yan, L.; Hu, L.; Park, S.; Ryu, M.; Zhou, X. Bioremediation of Petroleum Hydrocarbons Using Acinetobacter sp. SCYY-5 Isolated from Contaminated Oil Sludge: Strategy and Effectiveness Study. Int. J. Environ. Res. Public Health 2021, 18, 819. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Rehman, F.; Rafeeq, H.; Waqas, M.; Asghar, A.; Afsheen, N.; Rahdar, A.; Bilal, M.; Iqbal, H.M.N. In-situ, Ex-situ, and nano-remediation strategies to treat polluted soil, water, and air—A review. Chemosphere 2022, 289, 133252. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, K.S.; Puustinen, J.; Suortti, A.-M. Bioremediation of petroleum hydrocarbon-contaminated soil by composting in biopiles. Environ. Pollut. 2000, 107, 245–254. [Google Scholar] [CrossRef]
- Benyahia, F.; Embaby, A.S. Bioremediation of Crude Oil Contaminated Desert Soil: Effect of Biostimulation, Bioaugmentation and Bioavailability in Biopile Treatment Systems. Int. J. Environ. Res. Public Health 2016, 13, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steliga, T.; Kluk, D. Assessment of the Suitability of Melilotus officinalis for Phytoremediation of Soil Contaminated with Petroleum Hydrocarbons (TPH and PAH), Zn, Pb and Cd Based on Toxicological Tests. Toxics 2021, 9, 148. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum Hydrocarbon-Degrading Bacteria for the Remediation of Oil Pollution Under Aerobic Conditions: A Perspective Analysis. Front. Microbiol. 2018, 9, 2885. [Google Scholar] [CrossRef]
- Jariyal, M.; Yadav, M.; Singh, N.K.; Yadav, S.; Sharma, I.; Dahiya, S.; Thanki, A. 8—Microbial remediation progress and future prospects. In Bioremediation of Pollutants; Pandey, V.C., Singh, V., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 187–214. [Google Scholar]
- Sharma, B.; Shukla, P. Designing synthetic microbial communities for effectual bioremediation: A review. Biocatal. Biotransform. 2020, 38, 405–414. [Google Scholar] [CrossRef]
- Rylott, E.L.; Bruce, N.C. How synthetic biology can help bioremediation. Curr. Opin. Chem. Biol. 2020, 58, 86–95. [Google Scholar] [CrossRef]
- Xin, F.; Dong, W.; Dai, Z.; Jiang, Y.; Yan, W.; Lv, Z.; Fang, Y.; Jiang, M. Chapter 9—Biosynthetic Technology and Bioprocess Engineering. In Current Developments in Biotechnology and Bioengineering; Singh, S.P., Pandey, A., Du, G., Kumar, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 207–232. [Google Scholar]
- Bhattacharjee, G.; Gohil, N.; Singh, V. 14—Synthetic biology approaches for bioremediation. In Bioremediation of Pollutants; Pandey, V.C., Singh, V., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 303–312. [Google Scholar]
- Meng, L.; Bao, M.; Sun, P. Construction of long-chain alkane degrading bacteria and its application in bioremediation of crude oil pollution. Int. J. Biol. Macromol. 2018, 119, 524–532. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, B.; Lan, Y.; Ma, T. Enhanced degradation of different crude oils by defined engineered consortia of Acinetobacter venetianus RAG-1 mutants based on their alkane metabolism. Bioresour. Technol. 2021, 327, 124787. [Google Scholar] [CrossRef] [PubMed]
- Samin, G.; Pavlova, M.; Arif, M.I.; Postema, C.P.; Damborsky, J.; Janssen, D.B. A Pseudomonas putida strain genetically engineered for 1,2,3-trichloropropane bioremediation. Appl. Environ. Microbiol. 2014, 80, 5467–5476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Yu, F.; Wang, Q.; Gu, X.; Chen, W. Cloning of Catechol 2,3-Dioxygenase Gene and Construction of a Stable Genetically Engineered Strain for Degrading Crude Oil. Indian J. Microbiol. 2014, 54, 59–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Salvador, M.; Saunders, E.; González, J.; Avignone-Rossa, C.; Jiménez, J.I. Properties of alternative microbial hosts used in synthetic biology: Towards the design of a modular chassis. Essays Biochem. 2016, 60, 303–313. [Google Scholar] [PubMed] [Green Version]
- Chi, H.; Wang, X.; Shao, Y.; Qin, Y.; Deng, Z.; Wang, L.; Chen, S. Engineering and modification of microbial chassis for systems and synthetic biology. Synth. Syst. Biotechnol. 2019, 4, 25–33. [Google Scholar] [CrossRef] [PubMed]
- McCarty, N.S.; Ledesma-Amaro, R. Synthetic Biology Tools to Engineer Microbial Communities for Biotechnology. Trends Biotechnol. 2019, 37, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Göpfrich, K.; Platzman, I.; Spatz, J.P. Mastering Complexity: Towards Bottom-up Construction of Multifunctional Eukaryotic Synthetic Cells. Trends Biotechnol. 2018, 36, 938–951. [Google Scholar] [CrossRef] [Green Version]
- Mutschler, H.; Robinson, T.; Tang, T.-D.; Wegner, S. Special Issue on Bottom-Up Synthetic Biology. ChemBioChem 2019, 20, 2533–2534. [Google Scholar] [CrossRef]
- Roberts, M.A.J.; Cranenburgh, R.M.; Stevens, M.P.; Oyston, P.C.F. Synthetic biology: Biology by design. Microbiology 2013, 159, 1219–1220. [Google Scholar] [CrossRef]
- Varjani, S.J.; Upasani, V.N. A new look on factors affecting microbial degradation of petroleum hydrocarbon pollutants. Int. Biodeterior. Biodegrad. 2017, 120, 71–83. [Google Scholar] [CrossRef]
- Hussain, I.; Aleti, G.; Naidu, R.; Puschenreiter, M.; Mahmood, Q.; Rahman, M.M.; Wang, F.; Shaheen, S.; Syed, J.H.; Reichenauer, T.G. Microbe and plant assisted-remediation of organic xenobiotics and its enhancement by genetically modified organisms and recombinant technology: A review. Sci. Total Environ. 2018, 628–629, 1582–1599. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.M.; Lee, H.; Thai, T.D.; Shen, J.; Eyun, S.; Na, D. Synthetically engineered microbial scavengers for enhanced bioremediation. J. Hazard. Mater. 2021, 419, 126516. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, D.K.; Kim, J. New insights into bioremediation strategies for oil-contaminated soil in cold environments. Int. Biodeterior. Biodegrad. 2019, 142, 58–72. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, R.; Kumar, A.; Rai, R.; Singh, V.; Bhadouria, R. Genetically engineered bacteria for the degradation of dye and other organic compounds. In Abatement of Environmental Pollutants; Singh, P., Kumar, A., Borthakur, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 331–350. [Google Scholar]
- Pant, G.; Garlapati, D.; Agrawal, U.; Prasuna, R.G.; Mathimani, T.; Pugazhendhi, A. Biological approaches practised using genetically engineered microbes for a sustainable environment: A review. J. Hazard. Mater. 2021, 405, 124631. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Bilal, M.; Duan, X.; Iqbal, H.M.N. Mitigation of environmental pollution by genetically engineered bacteria—Current challenges and future perspectives. Sci. Total Environ. 2019, 667, 444–454. [Google Scholar] [CrossRef]
- Miri, S.; Naghdi, M.; Rouissi, T.; Kaur Brar, S.; Martel, R. Recent biotechnological advances in petroleum hydrocarbons degradation under cold climate conditions: A review. Crit. Rev. Environ. Sci. Technol. 2019, 49, 553–586. [Google Scholar] [CrossRef]
- Boronin, A.M.; Kosheleva, I.A. The Role of Catabolic Plasmids in Biodegradation of Petroleum Hydrocarbons. In Current Environmental Issues and Challenges; Cao, G., Orrù, R., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 159–168. [Google Scholar]
- Bhatt, P.; Bhandari, G.; Bhatt, K.; Maithani, D.; Mishra, S.; Gangola, S.; Bhatt, R.; Huang, Y.; Chen, S. Plasmid-mediated catabolism for the removal of xenobiotics from the environment. J. Hazard. Mater. 2021, 420, 126618. [Google Scholar] [CrossRef]
- Emamalipour, M.; Seidi, K.; Zununi Vahed, S.; Jahanban-Esfahlan, A.; Jaymand, M.; Majdi, H.; Amoozgar, Z.; Chitkushev, L.T.; Javaheri, T.; Jahanban-Esfahlan, R.; et al. Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression. Front. Cell Dev. Biol. 2020, 8, 229. [Google Scholar] [CrossRef]
- Yutin, N. Horizontal Gene Transfer. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 530–532. [Google Scholar]
- Abbasian, F.; Lockington, R.; Megharaj, M.; Naidu, R. A Review on the Genetics of Aliphatic and Aromatic Hydrocarbon Degradation. Appl. Biochem. Biotechnol. 2016, 178, 224–250. [Google Scholar] [CrossRef]
- Wang, B.; Xu, J.; Gao, J.; Fu, X.; Han, H.; Li, Z.; Wang, L.; Tian, Y.; Peng, R.; Yao, Q. Construction of an Escherichia coli strain to degrade phenol completely with two modified metabolic modules. J. Hazard. Mater. 2019, 373, 29–38. [Google Scholar] [CrossRef]
- Jones, J.A.; Koffas, M.A.G. Chapter Eight—Optimizing Metabolic Pathways for the Improved Production of Natural Products. Meth. Enzymol. 2016, 575, 179–193. [Google Scholar]
- Jain, C.K.; Gupta, M.; Prasad, Y.; Wadhwa, G.; Sharma, S.K. Homology modeling and protein engineering of alkane monooxygenase in Burkholderia thailandensis MSMB121: In silico insights. J. Mol. Model. 2014, 20, 2340–2343. [Google Scholar] [CrossRef] [PubMed]
- Dueber, J.E.; Wu, G.C.; Malmirchegini, G.R.; Moon, T.S.; Petzold, C.J.; Ullal, A.V.; Prather, K.L.J.; Keasling, J.D. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 2009, 27, 753–759. [Google Scholar] [CrossRef]
- Jiang, B.; Song, Y.; Liu, Z.; Huang, W.E.; Li, G.; Deng, S.; Xing, Y.; Zhang, D. Whole-cell bioreporters for evaluating petroleum hydrocarbon contamination. Crit. Rev. Environ. Sci. Technol. 2021, 51, 272–322. [Google Scholar] [CrossRef]
- Behera, B.K.; Das, A.; Sarkar, D.J.; Weerathunge, P.; Parida, P.K.; Das, B.K.; Thavamani, P.; Ramanathan, R.; Bansal, V. Polycyclic Aromatic Hydrocarbons (PAHs) in inland aquatic ecosystems: Perils and remedies through biosensors and bioremediation. Environ. Pollut. 2018, 241, 212–233. [Google Scholar] [CrossRef] [PubMed]
- Nandimandalam, H.; Gude, V.G. Indigenous biosensors for in situ hydrocarbon detection in aquatic environments. Mar. Pollut. Bull. 2019, 149, 110643. [Google Scholar] [CrossRef]
- Paitan, Y.; Biran, I.; Shechter, N.; Biran, D.; Rishpon, J.; Ron, E.Z. Monitoring aromatic hydrocarbons by whole cell electrochemical biosensors. Anal. Biochem. 2004, 335, 175–183. [Google Scholar] [CrossRef]
- Rajkumar, P.; Ramprasath, T.; Selvam, G.S. 12—A simple whole cell microbial biosensors to monitor soil pollution. In New Pesticides and Soil Sensors; Grumezescu, A.M., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 437–481. [Google Scholar]
- Nigam, V.K.; Pratyoosh, S. Enzyme Based Biosensors for Detection of Environmental Pollutants-A Review. J. Microbiol. Biotechnol. 2015, 25, 1773–1781. [Google Scholar] [CrossRef]
- Voon, C.H.; Yusop, N.M.; Khor, S.M. The state-of-the-art in bioluminescent whole-cell biosensor technology for detecting various organic compounds in oil and grease content in wastewater: From the lab to the field. Talanta 2022, 241, 123271. [Google Scholar] [CrossRef]
- Moratti, C.F.; Scott, C.; Coleman, N.V. Synthetic Biology Approaches to Hydrocarbon Biosensors: A Review. Front. Bioeng. Biotechnol. 2022, 9, 804234. [Google Scholar] [CrossRef]
- Tecon, R.; Van der Meer, J.R. Bacterial Biosensors for Measuring Availability of Environmental Pollutants. Sensors 2008, 8, 4062–4080. [Google Scholar] [CrossRef] [Green Version]
- Plotnikova, E.G.; Shumkova, E.S.; Shumkov, M.S. Whole-cell bacterial biosensors for the detection of aromatic hydrocarbons and their chlorinated derivatives. Appl. Biochem. Microbiol. 2016, 52, 347–357. [Google Scholar] [CrossRef]
- Patel, R.; Zaveri, P.; Mukherjee, A.; Agarwal, P.K.; More, P.; Munshi, N.S. Development of fluorescent protein-based biosensing strains: A new tool for the detection of aromatic hydrocarbon pollutants in the environment. Ecotoxicol. Environ. Saf. 2019, 182, 109450. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Ray, S.; Chowdhury, A.; Anand, R. Tunable Multiplexed Whole-Cell Biosensors as Environmental Diagnostics for ppb-Level Detection of Aromatic Pollutants. ACS Sens. 2021, 6, 1933–1939. [Google Scholar] [CrossRef] [PubMed]
- Padmaperuma, G.; Butler, T.; Shuhaili, F.; Almalki, W.; Vaidyanathan, S. Microbial consortia: Concept and application in fruit crop management. In Fruit Crops; Elsevier: Amsterdam, The Netherlands, 2020; pp. 353–366. [Google Scholar]
- Khoo, K.S.; Chew, K.W.; Yew, G.Y.; Leong, W.H.; Chai, Y.H.; Show, P.L.; Chen, W. Recent advances in downstream processing of microalgae lipid recovery for biofuel production. Bioresour. Technol. 2020, 304, 122996. [Google Scholar] [CrossRef]
- Kong, W.; Meldgin, D.R.; Collins, J.J.; Lu, T. Designing microbial consortia with defined social interactions. Nat. Chem. Biol. 2018, 14, 821–829. [Google Scholar] [CrossRef]
- Zhang, Z.; Yan, C.; Zhang, H. Mutualism between antagonists: Its ecological and evolutionary implications. Integr. Zool. 2021, 16, 84–96. [Google Scholar] [CrossRef]
- Drew, G.C.; Stevens, E.J.; King, K.C. Microbial evolution and transitions along the parasite–mutualist continuum. Nat. Rev. Microbiol. 2021, 19, 623–638. [Google Scholar] [CrossRef]
- Bidja Abena, M.T.; Sodbaatar, N.; Li, T.; Damdinsuren, N.; Choidash, B.; Zhong, W. Crude Oil Biodegradation by Newly Isolated Bacterial Strains and Their Consortium Under Soil Microcosm Experiment. Appl. Biochem. Biotechnol. 2019, 189, 1223–1244. [Google Scholar] [CrossRef]
- Qian, X.; Chen, L.; Sui, Y.; Chen, C.; Zhang, W.; Zhou, J.; Dong, W.; Jiang, M.; Xin, F.; Ochsenreither, K. Biotechnological potential and applications of microbial consortia. Biotechnol. Adv. 2020, 40, 107500. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Singh, R.K.; Singh, D. Chapter 20—Microbe-based bioreactor system for bioremediation of organic contaminants: Present and future perspective. In Microbe Mediated Remediation of Environmental Contaminants; Kumar, A., Singh, V.K., Singh, P., Mishra, V.K., Eds.; Woodhead Publishing: Sawston, UK, 2021; pp. 241–253. [Google Scholar]
- Chen, W.; Kong, Y.; Li, J.; Sun, Y.; Min, J.; Hu, X. Enhanced biodegradation of crude oil by constructed bacterial consortium comprising salt-tolerant petroleum degraders and biosurfactant producers. Int. Biodeterior. Biodegrad. 2020, 154, 105047. [Google Scholar] [CrossRef]
- Guo, G.; Tian, F.; Ding, K.; Wang, L.; Liu, T.; Yang, F. Effect of a bacterial consortium on the degradation of polycyclic aromatic hydrocarbons and bacterial community composition in Chinese soils. Int. Biodeterior. Biodegrad. 2017, 123, 56–62. [Google Scholar] [CrossRef]
- Xu, C.; Yu, H. Insights into constructing a stable and efficient microbial consortium. Chin. J. Chem. Eng. 2021, 30, 112–120. [Google Scholar] [CrossRef]
- Jia, X.; He, Y.; Jiang, D.; Liu, C.; Lu, W. Construction and analysis of an engineered Escherichia coli-Pseudomonas aeruginosa co-culture consortium for phenanthrene bioremoval. Biochem. Eng. J. 2019, 148, 214–223. [Google Scholar] [CrossRef]
- Wu, C.; Li, F.; Yi, S.; Ge, F. Genetically engineered microbial remediation of soils co-contaminated by heavy metals and polycyclic aromatic hydrocarbons: Advances and ecological risk assessment. J. Environ. Manag. 2021, 296, 113185. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, W. Synthetic biology: Recent progress, biosafety and biosecurity concerns, and possible solutions. J. Biosaf. Biosecurity 2019, 1, 22–30. [Google Scholar] [CrossRef]
- de Lorenzo, V. Environmental biosafety in the age of Synthetic Biology: Do we really need a radical new approach? Bioessays 2010, 32, 926–931. [Google Scholar] [CrossRef]
- Cases, I.; de Lorenzo, V. Genetically modified organisms for the environment: Stories of success and failure and what we have learned from them. Int Microbiol. 2005, 8, 213–222. [Google Scholar]
- Torres, L.; Krüger, A.; Csibra, E.; Gianni, E.; Pinheiro, V.B. Synthetic biology approaches to biological containment: Pre-emptively tackling potential risks. Essays Biochem. 2016, 60, 393–410. [Google Scholar]
- Kim, D.; Lee, J.W. Genetic Biocontainment Systems for the Safe Use of Engineered Microorganisms. Biotechnol. Bioprocess Eng. 2020, 25, 974–984. [Google Scholar] [CrossRef]
| Bioremediation Technique | Treatment | % of Degradation | Reference |
|---|---|---|---|
| Biostimulation: compounds such as nutrients, oxygen, biopolymers, biosurfactants or fertilizers are added, in order to enhance microbial activity [13,14,15]. | Mineral fertilizer with dolomite flour | Total petroleum hydrocarbon (TPH) decreased by 47% in 15 months. | [13] |
| Bioaugmentation: addition of autochthonous, exogenous or genetically engineered microorganisms with catabolic activity to the contamination site [14,15,16]. | Microbial inoculum with Alcanivorax as the dominant genus. | TPH was reduced by 41% in 63 days. | [16] |
| Acinetobacter sp. SCYY-5 | TPH was removed by 69.17% in 10 days. | [17] | |
| Biopile: contaminated soil is piled followed by the addition of nutrients and oxygen, which enhance degradation. A water system can be added or organic materials that acts as bulking agents [14,18,19]. | Biopile system with crude oil, nutrients and Amnite P-300 (consortium of 10 strains mainly belonging to the Pseudomonas genus) | TPH were reduced by 77% in 156 days. | [20] |
| Phytoremediation: process based on plants and their associated microorganisms to degrade, remove or immobilize toxic compounds from the environment [14,15]. | Melilotus officinalis | TPH by 42.2% and PAH by 49.9% in 6 months. | [21] |
| Genetically Engineered Microorganisms | Results | Reference |
|---|---|---|
| almA gene was inserted into two plasmids and transformed into Escherichia coli. | Total crude oil biodegradation was up to 32% and 50% in 72 h. | [28] |
| Enzyme consortium of three mutant alkane hydroxylases (alkMa, alkMb, and almA) belonging to Acinetobacter venetianus strain RAG-1. | Degradation of light crude oil by 88.65% [15.23% more than the wild type (WT)], viscous crude oil was degraded by 90.05% (21.65% more than WT), and high waxy crude oil by 60.52% (13.06% more than WT), in 10 days. | [29] |
| An improved dehalogenase gene (dhaA31) was cloned behind the constitutive dhlA promoter into Pseudomonas putida MC4. | 1,2,3-Trichloropropane was degraded by 97% in 48 days. The enzyme showed 36-foldhigher activity and 26-foldhigher catalytic efficiency than the WT enzyme. | [30] |
| A recombinant strain was constructed from the integration of catechol 2,3-dioxygenase in Acinetobacter sp. BS3. | The biodegradation rate of the oil concentrations was 80% in 28 days. | [31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Díaz, V.; Pedroza-Rodríguez, A.M.; Ramos-Monroy, O.; Castillo-Carvajal, L.C. Synthetic Biology: A New Era in Hydrocarbon Bioremediation. Processes 2022, 10, 712. https://doi.org/10.3390/pr10040712
Jiménez-Díaz V, Pedroza-Rodríguez AM, Ramos-Monroy O, Castillo-Carvajal LC. Synthetic Biology: A New Era in Hydrocarbon Bioremediation. Processes. 2022; 10(4):712. https://doi.org/10.3390/pr10040712
Chicago/Turabian StyleJiménez-Díaz, Valentina, Aura Marina Pedroza-Rodríguez, Oswaldo Ramos-Monroy, and Laura C. Castillo-Carvajal. 2022. "Synthetic Biology: A New Era in Hydrocarbon Bioremediation" Processes 10, no. 4: 712. https://doi.org/10.3390/pr10040712
APA StyleJiménez-Díaz, V., Pedroza-Rodríguez, A. M., Ramos-Monroy, O., & Castillo-Carvajal, L. C. (2022). Synthetic Biology: A New Era in Hydrocarbon Bioremediation. Processes, 10(4), 712. https://doi.org/10.3390/pr10040712






