Proteomics Analysis of Zygosaccharomyces mellis in Response to Sugar Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Honey Samples
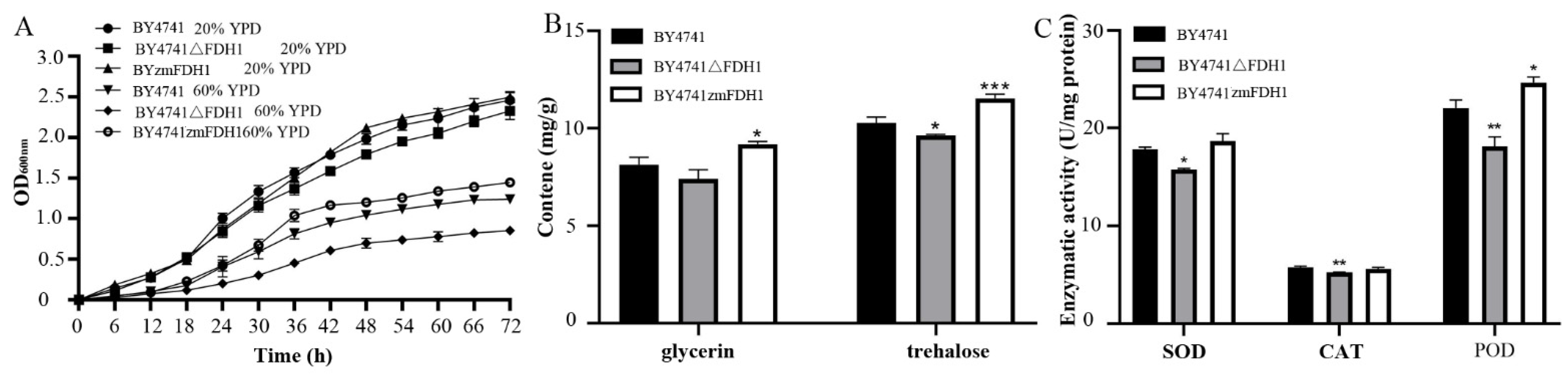
2.2. Growth Curve Measurements
2.3. POD, SOD and CAT Enzyme Activities Determination
2.4. Glycerol and Trehalose Content Determination
2.5. iTRAQ Experiment
2.6. Construction of the Mutant Strains
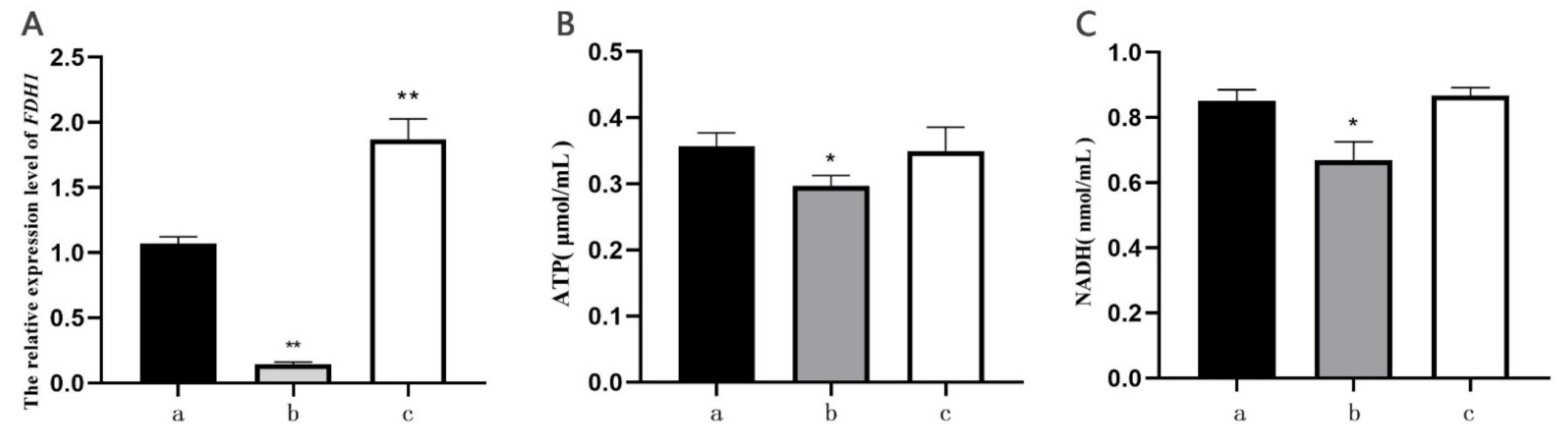
2.7. ATP and NADH Content Determination
2.8. qRT-PCR Amplification
2.9. Data Analysis
3. Results
3.1. Identification of Yeasts in Honey and the Influence of Sugar Stress on Yeast Growth
3.2. iTRAQ Data Analysis
3.3. Construction of the Mutant Strains Y4741△scFDH1 and BYzm4717FDH1
3.4. Effect of Sugar Stress on the Growth of BY4741△scFDH1 and BYzm4717FDH1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mateo, R.; Bosch-Reig, F. Sugar profiles of Spanish unifloral honeys. Food Chem. 1997, 60, 33–41. [Google Scholar] [CrossRef]
- Rawsthorne, H.; Phister, T.G. A real-time PCR assay for the enumeration and detection of Zygosaccharomyces bailii from wine and fruit juices. Int. J. Food Microbiol. 2006, 112, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.; Meirinho, S.; Estevinho, M.L.F.; Choupina, A. Yeast species associated with honey: Different identification methods. Arch. Zootec. 2010, 59, 103–113. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Tao, C.; Zhu, B.; Bai, W.; Zhang, L.; Wang, Z.; Liang, X. Identification of Zygosaccharomyces mellis strains in stored honey and their stress tolerance. Food Sci. Biotechnol. 2016, 25, 1645–1650. [Google Scholar] [CrossRef] [PubMed]
- Wrent, P.; Rivas, E.M.; Peinado, J.M.; de Silóniz, M.I. Strain typing of Zygosaccharomyces yeast species using a single molecular method based on polymorphism of the intergenic spacer region (IGS). Int. J. Food Microbiol. 2010, 142, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Bi, X.; Tao, C.; Fei, Y.; Gao, S.; Liang, J.; Bai, W. Comparative transcriptomics analysis of Zygosaccharomyces mellis under high-glucose stress. Food Sci. Hum. Wellness 2021, 10, 54–62. [Google Scholar] [CrossRef]
- Dakal, T.C.; Solieri, L.; Giudici, P. Adaptive response and tolerance to sugar and salt stress in the food yeast Zygosaccharomyces rouxii. Int. J. Food Microbiol. 2014, 185, 140–157. [Google Scholar] [CrossRef]
- Giardina, B.J.; Stanley, B.A.; Chiang, H.L. Comparative proteomic analysis of transition of Saccharomyces cerevisiae from glucose-deficient medium to glucose-rich medium. Proteome Sci. 2012, 10, 40. [Google Scholar] [CrossRef] [Green Version]
- Estéfani, G.R.; Amparo, Q.; José Manuel, G. iTRAQ-based proteome profiling of saccharomyces cerevisiae and cryotolerant species S. uvarum and S. kudriavzevii during low-temperature wine fermentation. J. Proteomics. 2016, 146, 70–79. [Google Scholar] [CrossRef]
- Saini, P.; Beniwal, A.; Kokkiligadda, A.; Vij, S. Response and tolerance of yeast to changing environmental stress during ethanol fermentation. Process Biochem. 2018, 72, 1–12. [Google Scholar] [CrossRef]
- Li, C.; Xu, Y.; Jiang, W.; Lv, X.; Dong, X. Effect of sodium chloride and cadmium on the growth, oxidative stress and antioxidant enzyme activities of Zygosaccharomyces rouxii. J. Ocean Univ. China 2014, 13, 460–466. [Google Scholar] [CrossRef]
- Saito, H.; Tatebayashi, K. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J. Biochem. 2004, 136, 267–272. [Google Scholar] [CrossRef]
- Babazadeh, R.; Furukawa, T.; Hohmann, S.; Furukawa, K. Rewiring yeast osmostress signalling through the MAPK network reveals essential and non-essential roles of Hog1 in osmoadaptation. Sci. Rep. 2014, 4, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Niu, C.; Liu, B.; Wei, J.P.; Wang, H.X.; Yuan, Y.H.; Yue, T.L. Protein abundance changes of Zygosaccharomyces rouxii in different sugar concentrations. Int. J. Food Microbiol. 2016, 233, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Noti, O.; Vaudano, E.; Giuffrida, M.G.; Lamberti, C.; Cavallarin, L.; Garcia-Moruno, E.; Pessione, E. Enhanced arginine biosynthesis and lower proteolytic profile as indicators of, Saccharomyces cerevisiae, stress in stationary phase during fermentation of high sugar grape must: A proteomic evidence. Food Res. Int. 2018, 105, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Põlme, S.; Riit, T.; Liiv, I.; Kõljalg, U.; Kisand, V.; Nilsson, R.H.; Hildebrand, F.; et al. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 2015, 10, 1–43. [Google Scholar] [CrossRef]
- Blaalid, R.; Kumar, S.; Nilsson, R.H.; Abarenkov, K.; Kirk, P.M.; Kauserud, H. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol. Mol. Ecol. Res. 2013, 13, 218–224. [Google Scholar] [CrossRef] [Green Version]
- Pompanon, F.; Coissac, E.; Taberlet, P. Metabarcoding, a new way of analysing biodiversity. Biofutur 2011, 319, 30–32. [Google Scholar]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bondioli, P.; Della, B.L. An alternative spectrophotometric method for the determination of free glycerol in biodiesel. Eur. J. Lipid Sci. Technol. 2005, 107, 153–157. [Google Scholar] [CrossRef]
- Wang, X.H.; Zhang, Y.Y.; Wang, C.; Wang, C.L.; Hou, L. Intracellular level of trehalose in soy sauce yeasts under different stresses. IERI Procedia 2013, 5, 321–326. [Google Scholar] [CrossRef] [Green Version]
- DiCarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stovicek, V.; Borodina, I.; Forster, J. CRISPR-Cas system enables fast and simple genome editing of industrial Saccharomyces cerevisiae strains. Metab. Eng. Commun. 2015, 2, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Stagos, D.; Soulitsiotis, N.; Tsadila, C.; Papaeconomou, S.; Arvanitis, C.; Ntontos, A.; Karkanta, F.; Adamou-Androulaki, S.; Petrotos, K.; Spandidos, D.A.; et al. Antibacterial and antioxidant activity of different types of honey derivedfrom Mount Olympus in Greece. Int. J. Mol. 2018, 42, 726–734. [Google Scholar] [CrossRef] [Green Version]
- Multari, S.; Guzzon, R.; Caruso, M.; Licciardello, C.; Martens, S. Alcoholic fermentation of citrus flavedo and albedo with pure and mixed yeast strains: Physicochemical characteristics and phytochemical profiles. LWT 2021, 144, 111133. [Google Scholar] [CrossRef]
- Benjaphokee, S.; Hasegawa, D.; Yokota, D.; Asvarak, T.; Auesukaree, C.; Sugiyama, M.; Kaneko, Y.; Boonchird, C.; Harashima, S. Highly efficient bioethanol production by a saccharomyces cerevisiae strain with multiple stress tolerance to high temperature, acid and ethanol. New Biotechnol. 2012, 29, 379–386. [Google Scholar] [CrossRef]
- Matallana, E.; Aranda, A. Biotechnological impact of stress response on wine yeast. Lett. Appl. Microbiol. 2017, 64, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Auesukaree, C. Molecular mechanisms of the yeast adaptive response and tolerance to stresses encountered during ethanol fermentation. J. Biosci. Bioeng. 2017, 124, 133–142. [Google Scholar] [CrossRef]
- Noti, O.; Vaudano, E.; Pessione, E.; Garcia-Moruno, E. Short-term response of different Saccharomyces cerevisiae strains to hyperosmotic stress caused by inoculation in grape must: RT-qPCR study and metabolite analysis. Food Microbiol. 2015, 52, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Petelenz-Kurdziel, E.; Kuehn, C.; Nordlander, B.; Klein, D.; Klipp, E. Quantitative analysis of glycerol accumulation, glycolysis and growth under osmotic stress. PLoS Comput. Biol. 2013, 9, e1003084. [Google Scholar] [CrossRef] [PubMed]
- Li, L.L.; Ye, Y.R.; Pan, L.; Zhu, Y.; Zheng, S.P.; Lin, Y. The induction of trehalose and glycerol in Saccharomyces cerevisiae in response to various stresses. Biochem. Biophys. Res. Commun. 2009, 387, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Willaert, R.G. The growth behavior of the model eukaryotic yeast Saccharomyces cerevisiae in microgravity. Curr. Biotechnol. 2013, 2, 226–234. [Google Scholar] [CrossRef]
- Hartland, R.P.; Vermeulen, C.A.; Sietsma, J.H.; Wessels, J.G.H.; Klis, F.M. The linkage of (1–3)-β-glucan to chitin during cell wall assembly in Saccharomyces cerevisiae. Yeast 1994, 10, 1591–1599. [Google Scholar] [CrossRef] [Green Version]
- Grün, C.H.; Hochstenbach, F.; Humbel, B.M.; Verkleij, A.J.; Sietsma, J.H.; Klis, F.M.; Kamerling, J.P.; Vliegenthart, J.F.G. The structure of cell wall α-glucan from fission yeast. Glycobiology 2005, 15, 245–257. [Google Scholar] [CrossRef] [Green Version]
- Vanegas, J.M.; Contreras, M.F.; Faller, R.; Longo, M.L. Role of unsaturated lipid and ergosterol in ethanol tolerance of model yeast biomembranes. Biophys. J. 2012, 102, 507–516. [Google Scholar] [CrossRef] [Green Version]
- Shobayashi, M.; Mitsueda, S.I.; Ago, M.; Fujii, T.; Iwashita, K.; Iefuji, H. Effects of culture conditions on ergosterol biosynthesis by Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2005, 69, 2381–2388. [Google Scholar] [CrossRef] [Green Version]
- Henderson, C.M.; Block, D.E. Examining the role of membrane lipid composition in determining the ethanol tolerance of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2014, 80, 2966–2972. [Google Scholar] [CrossRef] [Green Version]
- Mahmud, S.A.; Nagahisa, K.; Hirasawa, T.; Yoshikawa, K.; Ashitani, K.; Shimizu, H. Effect of trehalose accumulation on response to saline stress in Saccharomyces cerevisiae. Yeast 2009, 26, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Yoshiyama, Y.; Tanaka, K.; Yoshiyama, K.; Hibi, M.; Ogawa, J.; Shima, J. Trehalose accumulation enhances tolerance of Saccharomyces cerevisiae to acetic acid. J. Biosci. Bioeng. 2015, 119, 172–175. [Google Scholar] [CrossRef]
- Terrazas, W.D.M.; Aizemberg, R.; Gattas, E.A.D.L. Using Pichia pastoris to produce recombinant glycerol kinase. Rev. Cienc. Farm. Basica Apl. 2014, 35, 279–284. [Google Scholar]
- Wang, Z.X.; Kayingo, G.; Blomberg, A.; Prior, B.A. Cloning, sequencing and characterization of a gene encoding dihydroxyacetone kinase from Zygosaccharomyces rouxii NRRL2547. Yeast 2002, 19, 1447–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zufferey, R.; Mamoun, C.B. The initial step of glycerolipid metabolism in Leishmania major promastigotes involves a single glycerol-3-phosphate acyltransferase enzyme important for the synthesis of triacylglycerol but not essential for virulence. Mol. Microbiol. 2005, 56, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Myers, D.K.; Lawlor, D.T.; Attfield, P.V. Influence of invertase activity and glycerol synthesis and retention on fermentation of media with a high sugar concentration by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1997, 63, 145–150. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.Q.; Ren, L.; Zhang, J.; Reed, B.M.; Zhang, D.; Shen, X.H. Cryopreservation affects ROS-induced oxidative stress and antioxidant response in Arabidopsis seedlings. Cryobiology 2015, 70, 38–47. [Google Scholar] [CrossRef]
- Anjum, S.A.; Tanveer, M.; Hussain, S.; Shahzad, B.; Ashraf, U.; Fahad, S.; Tung, S.A. Osmoregulation and antioxidant production in maize under combined cadmium and arsenic stress. Environ. Sci. Pollut. Res. 2016, 23, 11864–11875. [Google Scholar] [CrossRef]
- Ferrareze, J.P.; Fugate, K.K.; Bolton, M.D.; Deckard, E.L.; Campbell, L.G.; Finger, F.L. Jasmonic acid does not increase oxidative defense mechanisms or common defense-related enzymes in postharvest sugarbeet roots. Postharvest Biol. Technol. 2013, 77, 11–18. [Google Scholar] [CrossRef]
- Lu, F.P.; Wang, Y.; Bai, D.Q.; Du, L.X. Adaptive response of Saccharomyces cerevisiae to hyperosmotic and oxidative stress. Process Biochem. 2005, 40, 3614–3618. [Google Scholar] [CrossRef]
- Sahu, S.; Das, P.; Ray, M.; Sabat, S.C. Osmolyte modulated enhanced rice leaf catalase activity under salt-stress. Adv. Biosci. Biotechnol. 2010, 1, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Das, A.B.; Sadowska-Bartosz, I.; Königstorfer, A.; Kettle, A.J.; Winterbourn, C.C. Superoxide dismutase protects ribonucleotide reductase from inactivation in yeast. Free Radic. Biol. Med. 2018, 116, 114–122. [Google Scholar] [CrossRef]
- Baroowa, B.; Gogoi, N. The effect of osmotic stress on anti-oxidative capacity of black gram (Vigna Mungo L.). Exp. Agric. 2017, 53, 84–99. [Google Scholar] [CrossRef] [Green Version]
- Haghdoost, N.S.; Salehi, T.Z.; Khosravi, A.; Sharifzadeh, A. Antifungal activity and influence of propolis against germ tube formation as a critical virulence attribute by clinical isolates of Candida albicans. J. Mycol Med. 2016, 26, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Ludovico, P.; Rodrigues, F.; Almeida, A.; Silva, M.T.; Barrientos, A.; Côrte-Real, M. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002, 13, 2598–2606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yale, J.; Bohnert, H.J. Transcript expression in Saccharomyces cerevisiae, at high salinity. J. Biol. Chem. 2001, 276, 15996–16007. [Google Scholar] [CrossRef] [Green Version]
- Tishkov, V.I.; Popov, V.O. Protein engineering of formate dehydrogenase. Biomol. Eng. 2006, 23, 89–110. [Google Scholar] [CrossRef]
- Suzuki, K.; Itai, R.; Suzuki, K.; Nakanishi, H.; Nishizawa, N.K.; Yoshimura, E.; Mori, Y.S. Formate dehydrogenase, an enzyme of anaerobic metabolism, is induced by iron deficiency in barley roots. Plant Physiol. 1998, 116, 725–732. [Google Scholar] [CrossRef] [Green Version]
- Hourton-Cabassa, C.; Ambard-Bretteville, F.; Moreau, F.; Davy de Virville, J.; Rémy, R.; Francs-Small, C.C. Stress Induction of Mitochondrial Formate Dehydrogenase in Potato Leaves. Plant Physiol. 1998, 116, 627–635. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Cai, R. Study and analytical application of inhibitory effect of captopril on multienzyme redox system. Talanta 2003, 61, 855–861. [Google Scholar] [CrossRef]
- Berríos-Rivera, S.J.; Bennett, G.N.; San, K.Y. Metabolic engineering of Escherichia coli: Increase of NADH availability by overexpressing an NAD(+)-dependent formate dehydrogenase. Metab. Eng. 2002, 4, 217–229. [Google Scholar] [CrossRef] [Green Version]
- Du, C.; Li, Y.; Xiang, R.; Yuan, W. Formate dehydrogenase improves the resistance to formic acid and acetic acid simultaneously in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2022, 23, 3406. [Google Scholar] [CrossRef]
- Liu, L.M.; Li, Y.; Shi, Z.P.; Du, G.C.; Chen, J. Enhancement of pyruvate productivity in torulopsis glabrata: Increase of NAD+ availability. J. Biotechnol. 2006, 126, 173–185. [Google Scholar] [CrossRef] [PubMed]

| Name | Sequences 5’ to 3’ |
|---|---|
| ITS1 | TCCGTAGGTGAACCTGCGG |
| ITS2 | GCTGCGTTCTTCATCGATGC |
| FDH1-UP-F | ATAGGCTTGAAAGAGAGTTTTAA |
| FDH1-UP-R | AATACTCATAATCACTCATTAATTTTCAGCTGTTATTTTG |
| FDH1-down-F | ATAACAGCTGAAAATTAATGAGTGATTATGAGTATTTGTGAGCA |
| FDH1-down-R | TTTTCTGCAGCAGATACTTTTGG |
| scFDH1-KpnI-F | GGGGTACCAACACAATGTCGAAGGGAAAGGTTTTGC |
| scFDH1-BamHI-R | CGGGATCCTTATTTCTTCTGTCCATAAGCTC |
| zmFDH1-KpnI-F | GGGGTACCAACACAATGTGCAAGGGCAAGGTGTC |
| zmFDH1-BamHI-R | CGGGATCCATGGTCAGAAAACAAACTAG |
| M13-F | CCCAGTCACGACGTTGTAAAACG |
| M13-R | AGCGGATAACAATTTCACACAGG |
| T7 | TAATACGACTCACTATAGGG |
| CYC1 Terminator | GTGACATAACTAATTACATGATG |
| FDH1-F | TTGCTGGCCTCGTTCAATCT |
| FDH1-R | CCGGCGTGGCTAAAAATGAG |
| Name | Relevant Description | References |
|---|---|---|
| Strains | ||
| BY4741 | S288C-derivative laboratory strain, MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 16 |
| E. coli DH5 α | F-, φ80, lacZΔM15, Δ(lacZYA-argF) U169 endA1, recA1, hsdR17(rk-, mk+) supE44, λ-, thi-1, gyrA96, relA1, phoA | Takara Bio, Kyoto, Japan |
| BY4741△FDH1 | Saccharomyces cerevisiae BY4741, △FDH1 | This study |
| BYzm4741FDH1 | BY4741△FDH1, zmFDH1 | This study |
| plasmid | ||
| Cas9 | p414-TEF1p-Cas9-CYC1t, CEN6/ARSH4 origin, TRP1, TEF1p promoter, codon optimized Cas9 with C-terminal SV40 tag, AmpR | Addgene |
| gRNA | p426-SNR52p-gRNA.CAN1.Y-SUP4t, PSNR52-gRNA-SUP4TT | Addgene |
| sgRNA-gRNA | Ligation sgRNA into the gRNA scaffold of plasmid gRNA | This study |
| ZmFDH1-PYES2 | Ligation zmFDH1 into the plasmid PYES2 | This study |
| Protein_ID | Description | 60% Glucose Group vs. 2% Glucose Group |
|---|---|---|
| TCA cycle | ||
| GCE97816.1 | NAD-dependent isocitrate dehydrogenase | 1.33 |
| GCF00601.1 | phosphoglycerate kinase | 0.74 |
| GCE97119.1 | cytochrome b subunit of succinate dehydrogenase, Sdh3p | 1.25 |
| GCE99324.1 | NADP-dependent isocitrate dehydrogenase | 0.44 |
| GCF01027.1 | malate dehydrogenase, cytoplasmic | 1.25 |
| GCE98908.1 | 2-oxoglutarate dehydrogenase complex E2 component | 1.28 |
| GCE99351.1 | citrate (Si)-synthase | 1.29 |
| GCF00374.1 | succinate dehydrogenase complex, subunit B | 1.58 |
| GCE97219.1 | succinate dehydrogenase assembly factor 2 | 0.79 |
| GCF01476.1 | membrane anchor subunit of succinate dehydrogenase, Sdh4 | 1.48 |
| Glycolysis | ||
| GCF00125.1 | glucokinase | 0.58 |
| GCF00259.1 | hexokinase A | 0.64 |
| GCE99269.1 | fructose-bisphosphate aldolase 1 | 0.69 |
| GCE98585.1 | glyceraldehyde-3-phosphate dehydrogenase | 1.33 |
| GCE97503.1 | glyceraldehyde-3-phosphate dehydrogenase 1 | 1.21 |
| Pentose phosphate pathway | ||
| GCF00070.1 | enolase-phosphatase E1 | 0.55 |
| GCE97493.1 | ribulose-phosphate 3-epimerase | 0.67 |
| GCF01076.1 | sedoheptulose-7-phosphate: D-glyceraldehyde-3-phosphate transaldolase | 0.69 |
| GCF01082.1 | glucose-6-phosphate isomerase | 0.78 |
| GCE98569.1 | triosephosphate isomerase | 0.8 |
| GCF01275.1 | glucose-6-phosphate 1-dehydrogenase | 1.25 |
| GCF00672.1 | ribose-5-phosphate isomerase rki1 | 1.55 |
| Respiratory chain | ||
| GCF00431.1 | ubiquinol-cytochrome c reductase core subunit 10 qcr10 | 1.4 |
| GCE98211.1 | NADH: ubiquinone oxidoreductase | 0.48 |
| GCF01526.1 | cytochrome c oxidase subunit 6 | 0.81 |
| GCE97076.1 | cytochrome c oxidase subunit 4 | 1.26 |
| GCF00071.1 | iso-1-cytochrome c | 1.29 |
| GCF01652.1 | cytochrome c oxidase subunit 2 | 1.3 |
| GCE99676.1 | Cu-binding protein | 1.31 |
| Trehalose and glycerol degradation | ||
| GCE99412.1 | phosphoglucomutase-2 | 0.62 |
| GCF00370.1 | glycerol kinase | 0.79 |
| GCE99841.1 | glycerol-3-phosphate/dihydroxyacetone phosphate acyltransferase | 0.83 |
| GCE98648.1 | dihydroxyacetone kinase 1 | 0.72 |
| Antioxidant | ||
| GCE98999.1 | superoxide dismutase (Mn), mitochondrial | 1.21 |
| GCE99208.1 | catalase T | 2 |
| Ergosterol synthesis | ||
| GCE98125.1 | phosphomevalonate kinase | 0.64 |
| GCE98064.1 | diphosphomevalonate decarboxylase | 0.68 |
| GCE98950.1 | phospholipid:diacylglycerol acyltransferase | 0.77 |
| GCF01680.1 | squalene epoxidase | 0.8 |
| GCE99731.1 | erg10, acetyl-CoA C-acetyltransferase | 1.61 |
| GCE99856.1 | C-24(28) sterol reductase | 1.49 |
| GCE99284.1 | RNA polymerase C-22 sterol desaturase | 1.52 |
| GCE97050.1 | farnesyl pyrophosphate synthetase | 0.66 |
| Cell wall | ||
| GCE99669.1 | UDP-glucose-4-epimerase | 0.48 |
| GCE98182.1 | UDP-N-acetylglucosamine pyrophosphorylase | 0.73 |
| GCE99673.1 | chitin synthase, class 3 | 0.75 |
| GCE99446.1 | glutamine-fructose-6-phosphate transaminase | 0.77 |
| GCE99630.1 | chitin synthase, class 1 | 0.82 |
| GCE97781.1 | Peroxin | 0.75 |
| NADH generation | ||
| GCE97061.1 | formate dehydrogenase 1 | 1.71 |
| GCE97099.1 | formate dehydrogenase 1 | 1.84 |
| GCE98400.1 | formate dehydrogenase 1 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Zhu, Y.; Li, Y.; Yang, W.; Zhou, H.; Chen, X. Proteomics Analysis of Zygosaccharomyces mellis in Response to Sugar Stress. Processes 2022, 10, 1193. https://doi.org/10.3390/pr10061193
Xu X, Zhu Y, Li Y, Yang W, Zhou H, Chen X. Proteomics Analysis of Zygosaccharomyces mellis in Response to Sugar Stress. Processes. 2022; 10(6):1193. https://doi.org/10.3390/pr10061193
Chicago/Turabian StyleXu, Xiaolan, Yuxuan Zhu, Yujie Li, Wenchao Yang, Hao Zhou, and Xinchao Chen. 2022. "Proteomics Analysis of Zygosaccharomyces mellis in Response to Sugar Stress" Processes 10, no. 6: 1193. https://doi.org/10.3390/pr10061193
APA StyleXu, X., Zhu, Y., Li, Y., Yang, W., Zhou, H., & Chen, X. (2022). Proteomics Analysis of Zygosaccharomyces mellis in Response to Sugar Stress. Processes, 10(6), 1193. https://doi.org/10.3390/pr10061193





