Multi-Models of Analyzing Dermoscopy Images for Early Detection of Multi-Class Skin Lesions Based on Fused Features
Abstract
:1. Introduction
- Removal of artifacts from dermoscopic images of skin lesions using an averaging filter and then inputting the images into a Laplacian filter to show the edges of the low-contrast lesions.
- Selection of important features and removal of redundant ones by PCA.
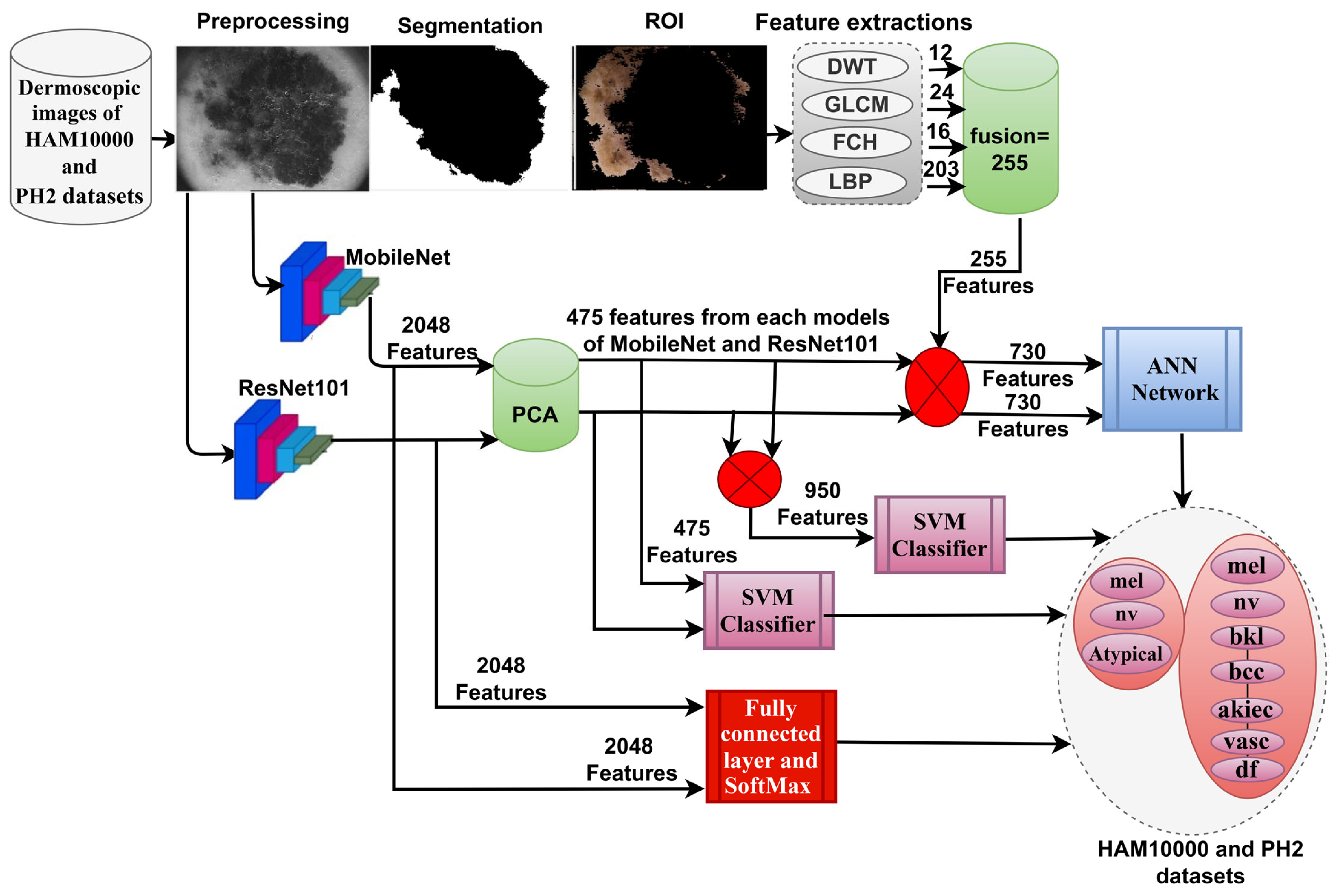
- Classification of dermoscopic images of two datasets, HAM10000 and PH2, by a hybrid technique, namely SVM-MobileNet-ResNet101.
- Classification of dermoscopic images of two datasets, HAM10000 and PH2, by ANN based on the combination of CNN (MobileNet and ResNet101) and handcrafted (discrete wavelet transform (DWT), gray-level co-occurrence matrix (GLCM), fuzzy color histogram (FCH) and local binary patterns (LBP)) features.
2. Related Work
3. Methods and Materials
3.1. Description of Dermoscopic Images Dataset
3.1.1. Description of HAM10000 Dataset
3.1.2. Description of PH2 Dataset
3.2. Pre-Processing Dermoscopic Images
3.2.1. Enhancement of Dermoscopic Images for the HAM10000 and PH2 Datasets
3.2.2. Hair Removal Method
3.3. SVM Based on CNN Features
3.3.1. Extract Deep Feature Maps
3.3.2. Machine Learning (SVM)
3.4. ANN with Hybrid Features of CNN and Handcrafted
4. Results of System Performance
4.1. Split of HAM10000 and PH2 Data Sets
4.2. Metrics of Systems Evaluation
4.3. Data Augmentation with Data Balancing
4.4. Results of Pre-Trained CNN Models
4.5. Results of SVM Based on CNN Features
4.6. Results of ANN with Hybrid Features of CNN and Handcrafted
5. Discussion the Results of the Systems
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bristow, R.E.; Chang, J.; Ziogas, A.; Campos, B.; Chavez, L.R. Anton-Culver, H. Impact of National Cancer Institute Comprehensive Cancer Centers on ovarian cancer treatment and survival. J. Am. Coll. Surg. 2015, 220, 940–950. Available online: https://www.sciencedirect.com/science/article/pii/S1072751515001258 (accessed on 15 November 2022). [CrossRef] [PubMed] [Green Version]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Kadampur, M.A.; Al Riyaee, S. Skin cancer detection: Applying a deep learning based model driven architecture in the cloud for classifying dermal cell images. Inform. Med. Unlocked 2020, 18, 100282. [Google Scholar] [CrossRef]
- Bilal, H.; Xiao, Y.; Khan, M.N.; Chen, J.; Wang, Q.; Zeng, Y.; Lin, X. Stabilization of Acne Vulgaris-Associated Microbial Dysbiosis with 2% Supramolecular Salicylic Acid. Pharmaceuticals 2023, 16, 87. [Google Scholar] [CrossRef]
- Singaporean Journal of Scientific Research (SJSR). Computer aided melanoma skin cancer detection using artificial neural network classifier. J. Sel. Areas Microelectron. 2016, 8, 35–42. Available online: http://www.sjsronline.com/Papers/Papers/sjsrvol8no22016-5.pdf (accessed on 22 December 2022).
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS ONE 2019, 14, e0217293. [Google Scholar] [CrossRef] [Green Version]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, M.E.; Menzies, S.W. Automated diagnostic instruments for cutaneous melanoma. In Seminars in Cutaneous Medicine and Surgery; WB Saunders: Philadelphia, PN, USA, 2008; Volume 27, pp. 32–36. [Google Scholar] [CrossRef]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Halpern, A. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging, Washington, DC, USA, 4–7 April 2018; IEEE: Piscatvie, NJ, USA, 2018; pp. 168–172. Available online: https://ieeexplore.ieee.org/abstract/document/8363547/ (accessed on 15 November 2022).
- Srinivasu, P.N.; SivaSai, J.G.; Ijaz, M.F.; Bhoi, A.K.; Kim, W.; Kang, J.J. Classification of Skin Disease Using Deep Learning Neural Networks with MobileNet V2 and LSTM. Sensors 2021, 21, 2852. [Google Scholar] [CrossRef] [PubMed]
- Alazzam, M.B.; Alassery, F.; Almulihi, A. Diagnosis of melanoma using deep learning. Math. Probl. Eng. 2021, 2021, 1423605. Available online: https://www.hindawi.com/journals/mpe/2021/1423605/ (accessed on 15 November 2022). [CrossRef]
- Jaworek-Korjakowska, J.; Brodzicki, A.; Cassidy, B.; Kendrick, C.; Yap, M.H. Interpretability of a Deep Learning Based Approach for the Classification of Skin Lesions into Main Anatomic Body Sites. Cancers 2021, 13, 6048. [Google Scholar] [CrossRef] [PubMed]
- Alam, T.M.; Shaukat, K.; Khan, W.A.; Hameed, I.A.; Almuqren, L.A.; Raza, M.A.; Aslam, M.; Luo, S. An Efficient Deep Learning-Based Skin Cancer Classifier for an Imbalanced Dataset. Diagnostics 2022, 12, 2115. [Google Scholar] [CrossRef] [PubMed]
- Abunadi, I.; Senan, E.M. Deep Learning and Machine Learning Techniques of Diagnosis Dermoscopy Images for Early Detection of Skin Diseases. Electronics 2021, 10, 3158. [Google Scholar] [CrossRef]
- Khamparia, A.; Singh, P.K.; Rani, P.; Samanta, D.; Khanna, A.; Bhushan, B. An internet of health things-driven deep learning framework for detection and classification of skin cancer using transfer learning. Trans. Emerg. Telecommun. Technol. 2021, 32, e3963. [Google Scholar] [CrossRef]
- Gouda, W.; Sama, N.U.; Al-Waakid, G.; Humayun, M.; Jhanjhi, N.Z. Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare 2022, 10, 1183. [Google Scholar] [CrossRef]
- Iqbal, I.; Younus, M.; Walayat, K.; Kakar, M.U.; Ma, J. Automated multi-class classification of skin lesions through deep convolutional neural network with dermoscopic images. Comput. Med. Imaging Graph. 2021, 88, 101843. [Google Scholar] [CrossRef]
- Murugan, A.; Nair, S.A.H.; Preethi, A.A.P.; Kumar, K.S. Diagnosis of skin cancer using machine learning techniques. Microprocess. Microsyst. 2021, 81, 103727. [Google Scholar] [CrossRef]
- Khan, M.A.; Sharif, M.; Akram, T.; Damaševičius, R.; Maskeliūnas, R. Skin Lesion Segmentation and Multiclass Classification Using Deep Learning Features and Improved Moth Flame Optimization. Diagnostics 2021, 11, 811. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Tsui, Y.Y.; Mandal, M. Skin Lesion Segmentation Using Deep Learning with Auxiliary Task. J. Imaging 2021, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Adegun, A.A.; Viriri, S.; Yousaf, M.H. A Probabilistic-Based Deep Learning Model for Skin Lesion Segmentation. Appl. Sci. 2021, 11, 3025. [Google Scholar] [CrossRef]
- Afza, F.; Sharif, M.; Khan, M.A.; Tariq, U.; Yong, H.-S.; Cha, J. Multiclass Skin Lesion Classification Using Hybrid Deep Features Selection and Extreme Learning Machine. Sensors 2022, 22, 799. [Google Scholar] [CrossRef]
- Tschandl, P.; Rinner, C.; Apalla, Z.; Argenziano, G.; Codella, N.; Halpern, A.; Kittler, H. Human–computer collaboration for skin cancer recognition. Nat. Med. 2020, 26, 1229–1234. [Google Scholar] [CrossRef]
- ADDI—Automatic Computer-Based Diagnosis System for Dermoscopy Images. Available online: https://www.fc.up.pt/addi/ph2%20database.html (accessed on 30 December 2022).
- Ahmed, I.A.; Senan, E.M.; Rassem, T.H.; Ali, M.A.; Shatnawi, H.S.A.; Alwazer, S.M.; Alshahrani, M. Eye Tracking-Based Diagnosis and Early Detection of Autism Spectrum Disorder Using Machine Learning and Deep Learning Techniques. Electronics 2022, 11, 530. [Google Scholar] [CrossRef]
- Lyakhov, P.A.; Lyakhova, U.A.; Nagornov, N.N. System for the Recognizing of Pigmented Skin Lesions with Fusion and Analysis of Heterogeneous Data Based on a Multimodal Neural Network. Cancers 2022, 14, 1819. [Google Scholar] [CrossRef] [PubMed]
- Fati, S.M.; Senan, E.M.; ElHakim, N. Deep and Hybrid Learning Technique for Early Detection of Tuberculosis Based on X-ray Images Using Feature Fusion. Appl. Sci. 2022, 12, 7092. [Google Scholar] [CrossRef]
- Al-Mekhlafi, Z.G.; Senan, E.M.; Rassem, T.H.; Mohammed, B.A.; Makbol, N.M.; Alanazi, A.A.; Ghaleb, F.A. Deep Learning and Machine Learning for Early Detection of Stroke and Haemorrhage. Comput. Mater. Contin. 2022, 72, 775–796. Available online: http://eprints.bournemouth.ac.uk/36721/ (accessed on 15 November 2022). [CrossRef]
- Fati, S.M.; Senan, E.M.; Azar, A.T. Hybrid and Deep Learning Approach for Early Diagnosis of Lower Gastrointestinal Diseases. Sensors 2022, 22, 4079. [Google Scholar] [CrossRef]
- Mohammed, B.A.; Senan, E.M.; Al-Mekhlafi, Z.G.; Rassem, T.H.; Makbol, N.M.; Alanazi, A.A.; Almurayziq, T.S.; Ghaleb, F.A.; Sallam, A.A. Multi-Method Diagnosis of CT Images for Rapid Detection of Intracranial Hemorrhages Based on Deep and Hybrid Learning. Electronics 2022, 11, 2460. [Google Scholar] [CrossRef]
- Mohammed, B.A.; Senan, E.M.; Rassem, T.H.; Makbol, N.M.; Alanazi, A.A.; Al-Mekhlafi, Z.G.; Almurayziq, T.S.; Ghaleb, F.A. Multi-Method Analysis of Medical Records and MRI Images for Early Diagnosis of Dementia and Alzheimer’s Disease Based on Deep Learning and Hybrid Methods. Electronics 2021, 10, 2860. [Google Scholar] [CrossRef]
- Al-Mekhlafi, Z.G.; Senan, E.M.; Mohammed, B.A.; Alazmi, M.; Alayba, A.M.; Alreshidi, A.; Alshahrani, M. Diagnosis of Histopathological Images to Distinguish Types of Malignant Lymphomas Using Hybrid Techniques Based on Fusion Features. Electronics 2022, 11, 2865. [Google Scholar] [CrossRef]
- Senan, E.M.; Jadhav, M.E. Diagnosis of dermoscopy images for the detection of skin lesions using SVM and KNN. In Proceedings of the Third International Conference on Sustainable Computing, Jaipur, India, 19–20 March 2021; Springer: Singapore, 2022; pp. 125–134. [Google Scholar] [CrossRef]
- Senan, E.M.; Jadhav, M.E.; Kadam, A. Classification of PH2 images for early detection of skin diseases. In Proceedings of the 2021 6th International Conference for Convergence in Technology, Mumbai, India, (I2CT), 2–4 April 2021; (I2CT). IEEE: Piscataway, NJ, USA, 2021; pp. 1–7. [Google Scholar] [CrossRef]
- Senan, E.M.; Jadhav, M.E. Techniques for the Detection of Skin Lesions in PH 2 Dermoscopy Images Using Local Binary Pattern (LBP). In Proceedings of the International Conference on Recent Trends in Image Processing and Pattern Recognition, Aurangabad, India, 3–4 January 2020; Springer: Singapore, 2020; pp. 14–25. [Google Scholar] [CrossRef]
- Senan, E.M.; Abunadi, I.; Jadhav, M.E.; Fati, S.M. Score and Correlation Coefficient-Based Feature Selection for Predicting Heart Failure Diagnosis by Using Machine Learning Algorithms. Comput. Math. Methods Med. 2021, 2021, 8500314. [Google Scholar] [CrossRef] [PubMed]
- Senan, E.M.; Mohammed, J.M.E.; Rassem, T.H.; Aljaloud, A.S.; Mohammed, B.A.; Al-Mekhlafi, Z.G. Early Diagnosis of Brain Tumour MRI Images Using Hybrid Techniques between Deep and Machine Learning. Comput. Math. Methods Med. 2022, 2022, 8330833. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, B.A.; Senan, E.M.; Al-Mekhlafi, Z.G.; Alazmi, M.; Alayba, A.M.; Alanazi, A.A.; Alreshidi, A.; Alshahrani, M. Hybrid Techniques for Diagnosis with WSIs for Early Detection of Cervical Cancer Based on Fusion Features. Appl. Sci. 2022, 12, 8836. [Google Scholar] [CrossRef]
- Fraiwan, M.; Faouri, E. On the Automatic Detection and Classification of Skin Cancer Using Deep Transfer Learning. Sensors 2022, 22, 4963. [Google Scholar] [CrossRef] [PubMed]















| Datasets | HAM10000 | PH2 | ||||
|---|---|---|---|---|---|---|
| Phase | 80% (80:20) | Testing 20% | 80% (80:20) | Testing 20% | ||
| Classes | Training (80%) | Validation (20%) | Training (80%) | Validation (20%) | ||
| Melanocytic nevi (nv) | 4291 | 1073 | 1341 | 51 | 13 | 16 |
| Atypical | - | - | - | 51 | 13 | 16 |
| Melanoma (mel) | 712 | 178 | 223 | 26 | 6 | 8 |
| Benign keratosis lesions (bkl) | 703 | 176 | 220 | - | - | - |
| Basal cell carcinoma (bcc) | 329 | 82 | 103 | - | - | - |
| Actinic keratoses (akiec) | 210 | 52 | 65 | - | - | - |
| Vascular (vasc) | 91 | 23 | 28 | - | - | - |
| Dermatofibroma (df) | 74 | 18 | 23 | - | - | - |
| Datasets | HAM10000 | PH2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phase | Training Dataset | Training Dataset | ||||||||
| Classes | nv | Mel | bkl | bcc | akiec | vasc | df | nv | atypical | mel |
| Bef-augm | 4291 | 712 | 703 | 329 | 210 | 91 | 74 | 51 | 51 | 26 |
| Aft-augm | 4291 | 4272 | 4218 | 4277 | 4200 | 4095 | 3700 | 1530 | 1530 | 1300 |
| Dataset | Type of Class | AUC % | Accuracy % | Sensitivity % | Precision % | Specificity % |
|---|---|---|---|---|---|---|
| HAM10000 | akiec | 92.5 | 87.7 | 88.4 | 90.5 | 99.6 |
| bcc | 93.4 | 89.3 | 89.3 | 88.5 | 98.7 | |
| bkl | 96.2 | 93.6 | 94.1 | 97.6 | 99.7 | |
| df | 94.6 | 91.3 | 90.8 | 100 | 99.6 | |
| mel | 96.1 | 96.9 | 97.2 | 64.5 | 93.2 | |
| nv | 95.7 | 91.5 | 90.6 | 98.4 | 97.4 | |
| vasc | 94.6 | 71.4 | 91.3 | 90.9 | 99.5 | |
| average ratio | 94.73 | 91.80 | 91.67 | 90.06 | 98.24 | |
| PH2 | atypical | 92.2 | 87.5 | 88.2 | 100 | 99.6 |
| mel | 94.5 | 100 | 99.7 | 72.7 | 91.4 | |
| nv | 91.9 | 87.5 | 88.4 | 93.3 | 95.8 | |
| average ratio | 92.87 | 90.00 | 92.10 | 88.67 | 95.60 |
| Dataset | Type of Class | AUC % | Accuracy % | Sensitivity % | Precision % | Specificity % |
|---|---|---|---|---|---|---|
| HAM10000 | akiec | 95.4 | 93.8 | 94.3 | 87.1 | 99.5 |
| bcc | 93.7 | 92.2 | 91.6 | 92.2 | 99.6 | |
| bkl | 96.6 | 93.2 | 93.4 | 96.2 | 99.5 | |
| df | 93.6 | 91.3 | 90.8 | 95.5 | 99.8 | |
| mel | 96.1 | 93.3 | 93.2 | 94.2 | 93.4 | |
| nv | 98.2 | 91.1 | 90.7 | 98.8 | 97.8 | |
| vasc | 91.5 | 71.4 | 71.2 | 57.1 | 98.6 | |
| average ratio | 95.01 | 91.40 | 89.31 | 88.73 | 98.31 | |
| PH2 | atypical | 94.9 | 93.8 | 94.1 | 100 | 99.6 |
| mel | 90.8 | 87.5 | 87.7 | 87.5 | 97.2 | |
| nv | 93.7 | 93.8 | 94.4 | 88.2 | 91.9 | |
| average ratio | 93.13 | 92.50 | 92.07 | 91.90 | 96.23 |
| Dataset | Type of Class | AUC % | Accuracy % | Sensitivity % | Precision % | Specificity % |
|---|---|---|---|---|---|---|
| HAM10000 | akiec | 92.1 | 84.6 | 85.4 | 87.3 | 99.5 |
| bcc | 97.6 | 91.3 | 90.8 | 93.1 | 99.6 | |
| bkl | 98.2 | 93.6 | 93.7 | 90.4 | 98.7 | |
| df | 76.4 | 65.2 | 65.4 | 34.9 | 99.2 | |
| mel | 96.4 | 89.7 | 90.3 | 95.2 | 99.1 | |
| nv | 99.1 | 97.8 | 98.2 | 98.3 | 97.4 | |
| vasc | 88.2 | 78.6 | 79.4 | 91.7 | 99.6 | |
| average ratio | 92.57 | 95.00 | 86.17 | 84.41 | 99.01 | |
| PH2 | atypical | 95.6 | 87.5 | 88.1 | 100 | 99.5 |
| mel | 96.1 | 100 | 99.7 | 88.9 | 97.2 | |
| nv | 97.8 | 100 | 99.5 | 94.1 | 96.4 | |
| average ratio | 96.50 | 95.00 | 95.77 | 94.33 | 97.70 |
| Dataset | Type of Class | AUC % | Accuracy % | Sensitivity % | Precision % | Specificity % |
|---|---|---|---|---|---|---|
| HAM10000 | akiec | 84.2 | 80 | 80.4 | 77.6 | 99.2 |
| bcc | 87.5 | 86.4 | 85.6 | 83.2 | 98.8 | |
| bkl | 94.3 | 95.5 | 94.7 | 88.6 | 97.9 | |
| df | 91.2 | 87 | 87.2 | 90.9 | 99.5 | |
| mel | 89.4 | 85.2 | 85.1 | 91.8 | 99.3 | |
| nv | 97.6 | 98.4 | 97.7 | 98.9 | 98.1 | |
| vasc | 86.2 | 67.9 | 68.2 | 65.5 | 98.8 | |
| average ratio | 90.06 | 94.80 | 85.56 | 85.21 | 98.80 | |
| PH2 | atypical | 98.1 | 100 | 99.7 | 94.1 | 96.2 |
| mel | 93.5 | 87.5 | 88.4 | 100 | 99.6 | |
| nv | 95.5 | 93.8 | 94.1 | 93.8 | 95.8 | |
| average ratio | 95.70 | 95.00 | 94.07 | 95.97 | 97.20 |
| Dataset | Type of Class | AUC % | Accuracy % | Sensitivity % | Precision % | Specificity % |
|---|---|---|---|---|---|---|
| HAM10000 | akiec | 98.4 | 90.8 | 90.5 | 89.4 | 99.5 |
| bcc | 99.1 | 91.3 | 91.2 | 89.5 | 99.1 | |
| bkl | 98.4 | 96.4 | 95.8 | 96.8 | 99.5 | |
| df | 98.6 | 87 | 87.3 | 87 | 99.7 | |
| mel | 98.1 | 93.3 | 93.2 | 94.4 | 98.8 | |
| nv | 98.9 | 98.9 | 98.7 | 98.9 | 98.6 | |
| vasc | 99.3 | 82.1 | 82.4 | 82.1 | 99.6 | |
| average ratio | 98.69 | 97.00 | 91.30 | 91.16 | 99.26 | |
| PH2 | atypical | 97.6 | 93.8 | 94.3 | 100 | 99.8 |
| mel | 98.4 | 100 | 99.5 | 100 | 99.5 | |
| nv | 97.9 | 100 | 99.8 | 94.1 | 96.4 | |
| average ratio | 97.97 | 97.50 | 97.87 | 98.03 | 98.57 |
| Dataset | Type of Class | AUC % | Accuracy % | Sensitivity % | Precision % | Specificity % |
|---|---|---|---|---|---|---|
| HAM10000 | akiec | 96.7 | 90.8 | 91.2 | 95.2 | 99.5 |
| bcc | 98.2 | 95.1 | 94.6 | 96.1 | 99.6 | |
| bkl | 96.6 | 97.3 | 96.7 | 98.2 | 99.5 | |
| df | 98.2 | 100 | 99.5 | 95.8 | 99.7 | |
| mel | 99.3 | 98.7 | 98.7 | 97.3 | 99.6 | |
| nv | 99.5 | 99.5 | 98.1 | 99.6 | 98.7 | |
| vasc | 94.2 | 82.1 | 82.4 | 71.9 | 99.4 | |
| average ratio | 97.53 | 98.40 | 94.46 | 93.44 | 99.43 | |
| PH2 | atypical | 100 | 100 | 100 | 100 | 100 |
| mel | 100 | 100 | 100 | 100 | 100 | |
| nv | 100 | 100 | 100 | 100 | 100 | |
| average ratio | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| Dataset | Type of Class | AUC % | Accuracy % | Sensitivity % | Precision % | Specificity % |
|---|---|---|---|---|---|---|
| HAM10000 | akiec | 99.1 | 89.2 | 89.2 | 90.6 | 99.6 |
| bcc | 99.6 | 91.3 | 91.4 | 92.2 | 99.5 | |
| bkl | 98.7 | 96.4 | 96.2 | 97.2 | 99.7 | |
| df | 99.4 | 87 | 87.4 | 87 | 100 | |
| mel | 98.7 | 94.2 | 93.8 | 96.3 | 99.5 | |
| nv | 98.9 | 99.6 | 99.5 | 98.9 | 98.2 | |
| vasc | 99.4 | 85.7 | 86.5 | 88.9 | 99.7 | |
| average ratio | 99.11 | 97.60 | 92.00 | 93.01 | 99.46 | |
| PH2 | atypical | 99.5 | 100 | 99.7 | 100 | 99.9 |
| mel | 99.7 | 100 | 99.6 | 88.9 | 97.4 | |
| nv | 99.4 | 93.8 | 94.4 | 100 | 99.5 | |
| average ratio | 99.53 | 97.50 | 97.90 | 96.30 | 98.93 |
| Datasets | Radiomic Features | Gradient and Validation Check | Cross-Entropy | Error Histogram | Regression% |
|---|---|---|---|---|---|
| HAM10000 | MobileNet with handcrafted | 0.0065521 at epoch 47 | 0.049723 at epoch 41 | −0.9382 to 0.9502 | 98.11 |
| ResNet101 with handcrafted | 0.009407 at epoch 44 | 0.057363 to epoch 38 | −0.9498 to 0.9499 | 97.88 | |
| PH2 | MobileNet with handcrafted | 0.00090003 at epoch 46 | 0.02453 at epoch 40 | −0.9062 to 0.9067 | 95 |
| ResNet101 with handcrafted | 0.026715 at epoch 23 | 0.015564 to epoch 17 | −0.9363 to 0.9387 | 97.04 |
| Datasets | Techniques | Features | akiec | bcc | Bkl | df | mel | nv | vasc | Atypical | Accuracy % | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HAM10000 | MobileNet | 87.7 | 89.3 | 93.6 | 91.3 | 96.9 | 91.5 | 71.5 | 91.8 | |||
| ResNet-101 | 93.8 | 92.2 | 93.2 | 91.3 | 93.3 | 91.3 | 71.4 | - | 91.4 | |||
| SVM | MobileNet-PCA | 84.6 | 91.3 | 93.6 | 65.2 | 89.7 | 97.8 | 78.6 | - | 95 | ||
| ResNet101-PCA | 80 | 86.4 | 95.5 | 87 | 85.2 | 98.4 | 67.9 | - | 94.8 | |||
| Fusion features | MobileNet with ResNet-101 | 90.8 | 91.3 | 96.4 | 87 | 93.3 | 98.9 | 82.1 | - | 97 | ||
| ANN | Fusion features | MobileNet and handcrafted | 90.8 | 95.1 | 97.5 | 100 | 98.7 | 99.5 | 82.1 | - | 98.4 | |
| ResNet-101 and handcrafted | 89.2 | 91.3 | 96.4 | 87 | 94.2 | 99.6 | 85.7 | - | 97.6 | |||
| PH2 | MobileNet | - | - | - | - | 100 | 87.5 | 87.5 | 90 | |||
| ResNet-101 | - | - | - | - | 87.5 | 93.8 | - | 93.8 | 92.5 | |||
| SVM | MobileNet-PCA | - | - | - | - | 100 | 100 | - | 87.5 | 95 | ||
| ResNet101-PCA | - | - | - | - | 87.5 | 93.8 | - | 100 | 95 | |||
| Fusion features | MobileNet with ResNet-101 | - | - | - | - | 100 | 100 | - | 93.8 | 97.5 | ||
| ANN | Fusion features | MobileNet and handcrafted | - | - | - | - | 100 | 100 | - | 100 | 100 | |
| ResNet-101 and handcrafted | - | - | - | - | 100 | 93.8 | - | 100 | 97.5 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, I.A.; Senan, E.M.; Shatnawi, H.S.A.; Alkhraisha, Z.M.; Al-Azzam, M.M.A. Multi-Models of Analyzing Dermoscopy Images for Early Detection of Multi-Class Skin Lesions Based on Fused Features. Processes 2023, 11, 910. https://doi.org/10.3390/pr11030910
Ahmed IA, Senan EM, Shatnawi HSA, Alkhraisha ZM, Al-Azzam MMA. Multi-Models of Analyzing Dermoscopy Images for Early Detection of Multi-Class Skin Lesions Based on Fused Features. Processes. 2023; 11(3):910. https://doi.org/10.3390/pr11030910
Chicago/Turabian StyleAhmed, Ibrahim Abdulrab, Ebrahim Mohammed Senan, Hamzeh Salameh Ahmad Shatnawi, Ziad Mohammad Alkhraisha, and Mamoun Mohammad Ali Al-Azzam. 2023. "Multi-Models of Analyzing Dermoscopy Images for Early Detection of Multi-Class Skin Lesions Based on Fused Features" Processes 11, no. 3: 910. https://doi.org/10.3390/pr11030910
APA StyleAhmed, I. A., Senan, E. M., Shatnawi, H. S. A., Alkhraisha, Z. M., & Al-Azzam, M. M. A. (2023). Multi-Models of Analyzing Dermoscopy Images for Early Detection of Multi-Class Skin Lesions Based on Fused Features. Processes, 11(3), 910. https://doi.org/10.3390/pr11030910











