New Refined Experimental Analysis of Fungal Growth in Degraded Bio-Based Materials
Abstract
:1. Introduction
2. Materials and Methods
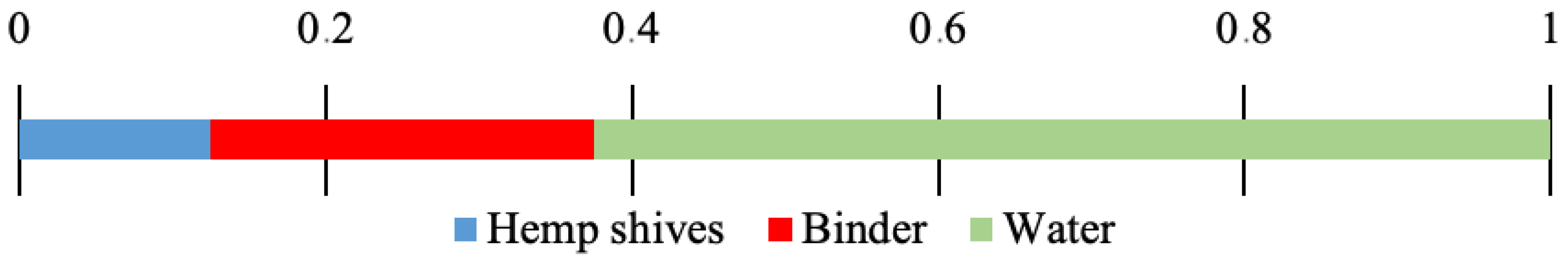
2.1. General Strategy of Experiment
2.2. In Situ Conditioning and Sample Preparation
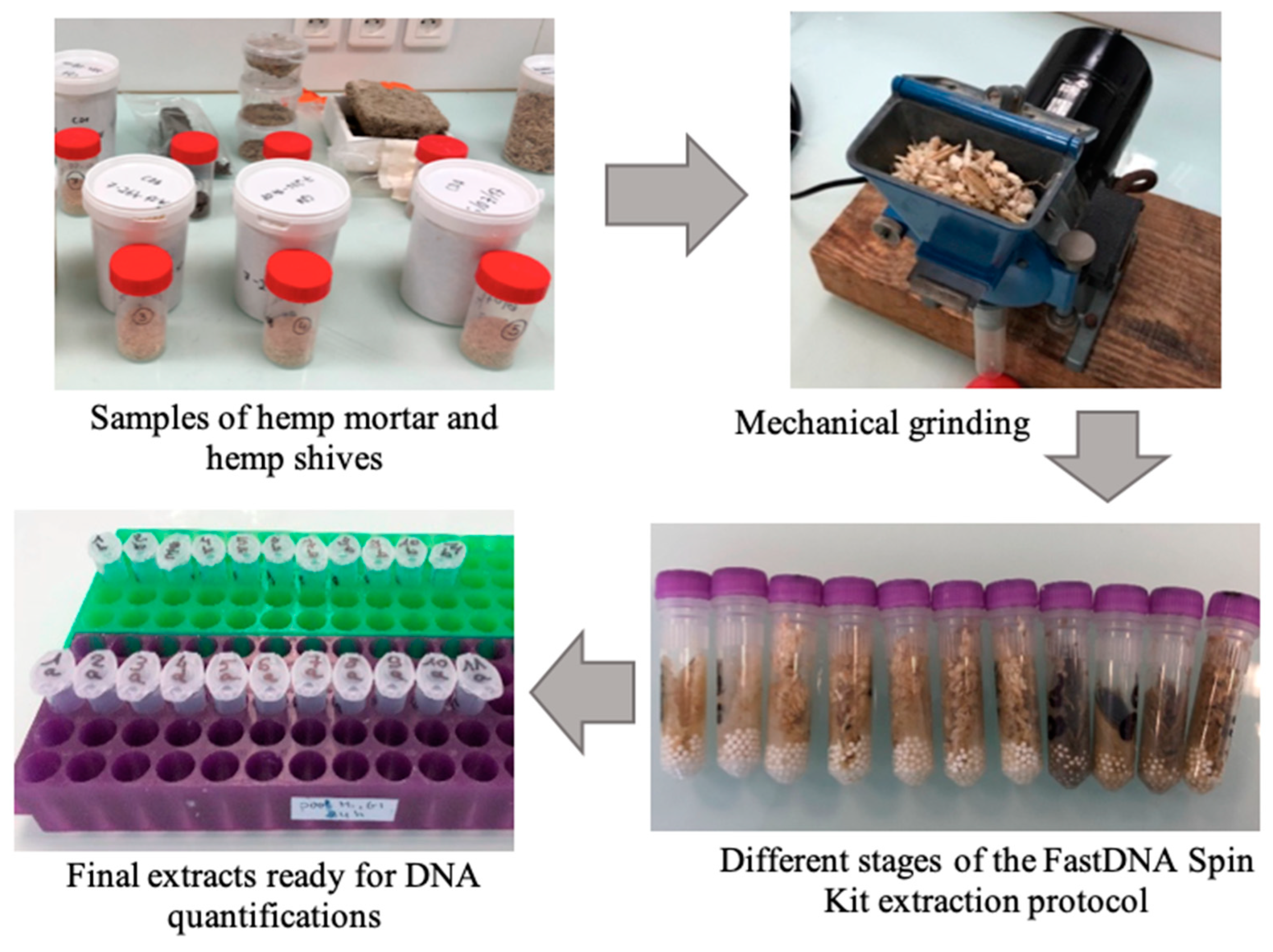
2.3. Analyses of Contaminated Hemp Mortar and Mold Proliferation
2.4. Metagenomic Analyses of Microbial Community
3. Results and Discussion
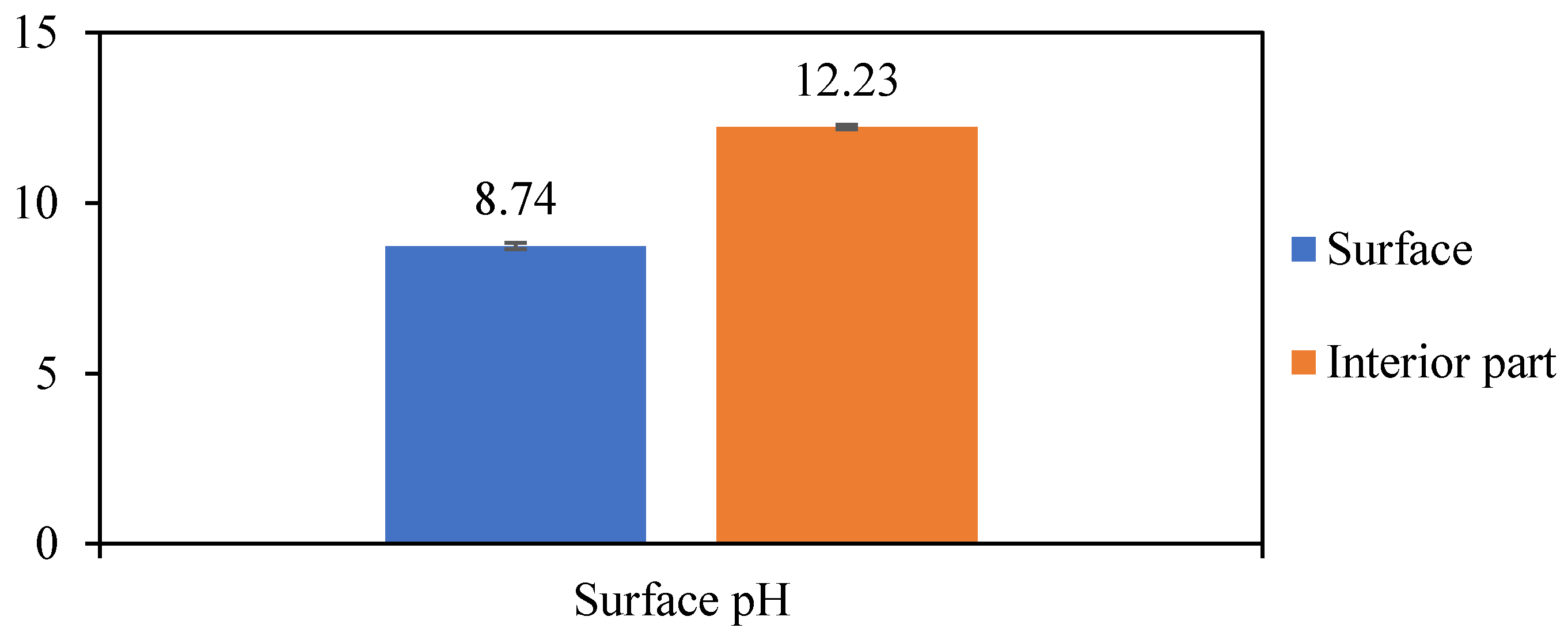
3.1. Hemp Mortar Surface Analysis
3.2. DNA Extraction
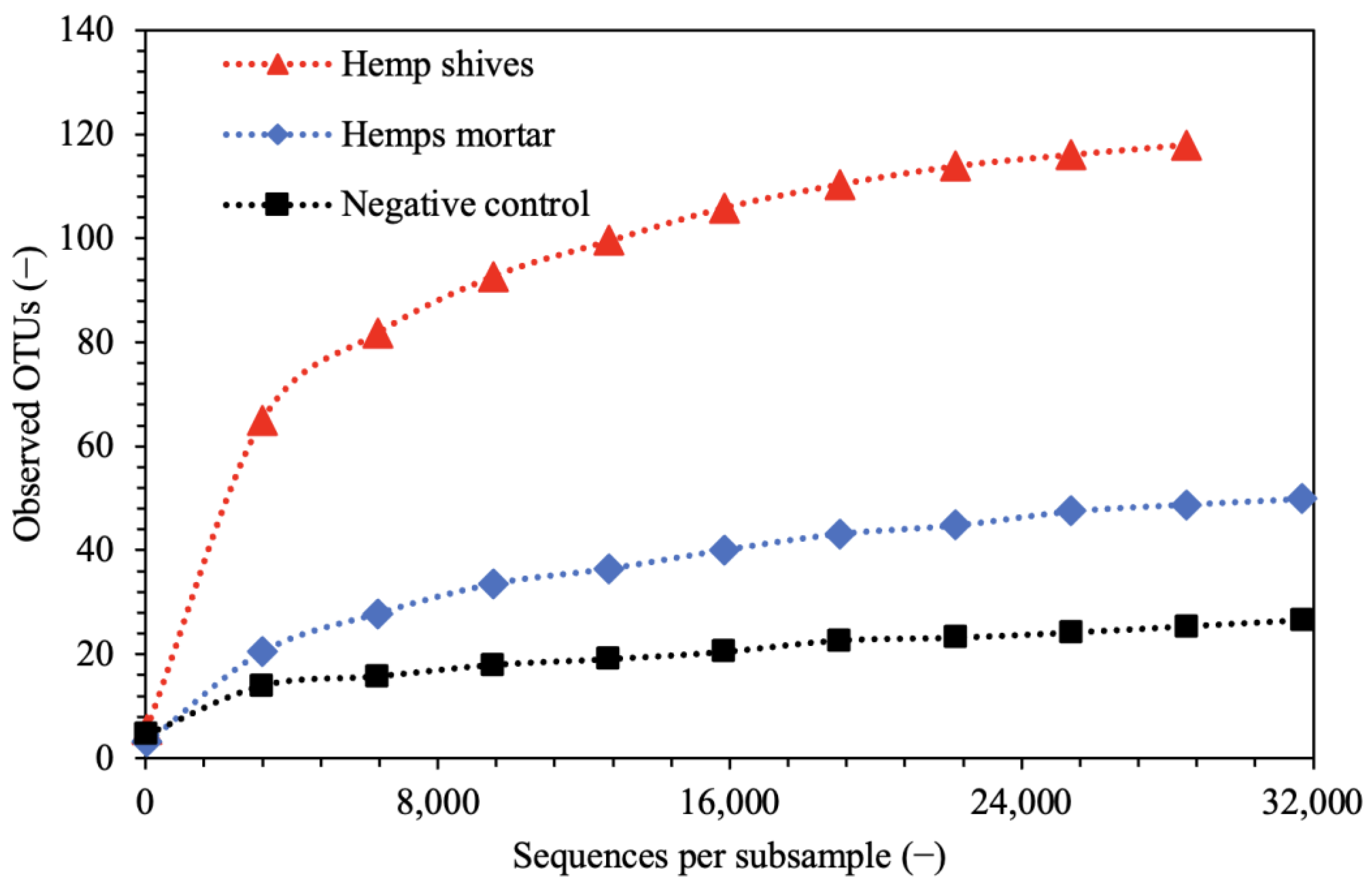
3.3. Rarefaction Analysis
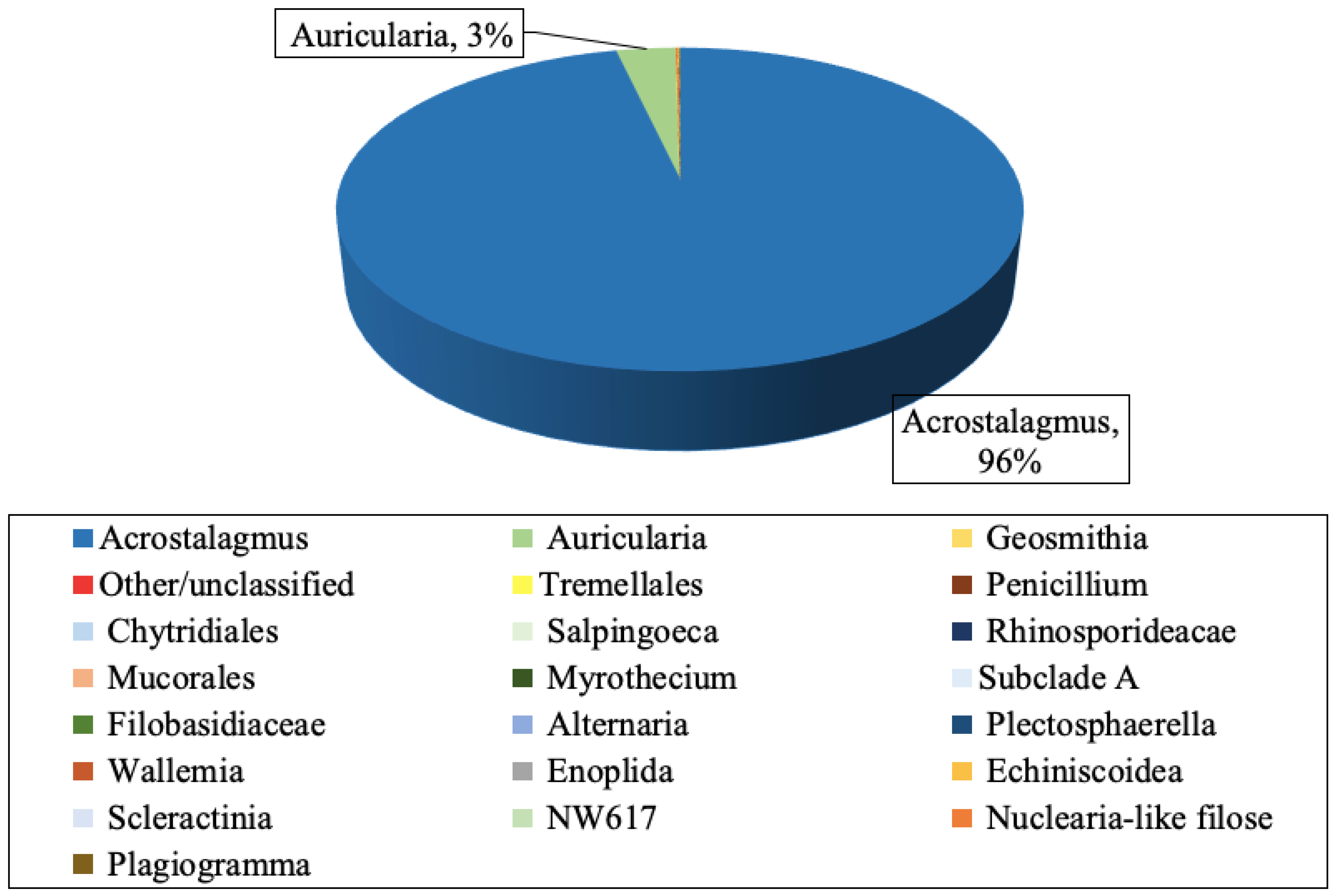
3.4. Mold Species Identified
4. Discussion
5. Conclusions
- -
- First, the surface of contaminated hemp mortar was analyzed with ATR-FTIR and W-SEM, which confirmed the presence of organics. Second, a DNA extraction method was carefully selected based on total and fungal extracted DNA quantification;
- -
- Then, the diversity of studied samples using rarefaction analysis was evaluated to ensure the representativeness of the results;
- -
- Finally, the mold genera present in hemp shives and hemp mortar were then accurately identified to analyze the trophic cycle of major fungi.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bennai, F.; Nabil, I.; Abahri, K.; Belarbi, R.; Tahakourt, A. Experimental characterization of thermal and hygric properties of hemp concrete with consideration of the material age evolution. Heat Mass Transfer. 2017, 54, 1189–1197. [Google Scholar] [CrossRef]
- Bennai, F.; El Hachem, C.; Abahri, K.; Belarbi, R. Microscopic hydric characterization of hemp concrete by X-ray microtomography and digital volume correlation. Constr. Build. Mater. 2018, 188, 983–994. [Google Scholar] [CrossRef]
- Rima, A.; Abahri, K.; Bennai, F.; El Hachem, C.; Bonnet, M. Microscopic estimation of swelling and shrinkage of hemp concrete in response to relative humidity variations. J. Build. Eng. 2021, 43, 102929. [Google Scholar] [CrossRef]
- Kosiachevskyi, D.; El Hachem, C.; Abahri, K.; Bennacer, R.; Chaouche, M. Biomaterials heterogeneous displacement, strain and swelling under hydric sorption/desorption: 2D image correlation on spruce wood. Constr. Build. Mater. 2021, 286, 122997. [Google Scholar] [CrossRef]
- Kosiachevskyi, D.; Abahri, K.; Daubresse, A.; Prat, E.; Chaouche, M. Analyse of the evolution of the hygrothermal properties of the hemp-based mortar within the framework of the total weathering protocol. Constr. Technol. Archit. 2022, 1, 35–42. [Google Scholar] [CrossRef]
- Walker, R.; Pavia, S.; Mitchell, R. Mechanical properties and durability of hemp-lime concretes. Constr. Build. Mater. 2014, 61, 340–348. [Google Scholar] [CrossRef]
- Sinka, M.; Obuka, V.; Bajare, D.; Jakovics, A. Durability and hygrothermal performance of bio-based materials in Northern European climate. Acad. J. Civ. Eng. 2019, 37, 371–377. [Google Scholar] [CrossRef]
- Watkinson, S.C.; Boddy, L.; Money, N.P. The Fungi, 3rd ed.; Elsevier, Ed.; Academic Press: Amsterdam, The Netherlands, 2015; p. 466. ISBN 9780123820341. [Google Scholar]
- Dallongeville, A.; Le Cann, P.; Zmirou-Navier, D.; Chevrier, C.; Costet, N.; Annesi-Maesano, I.; Blanchard, O. Concentration and determinants of molds and allergens in indoor air and house dust of French dwellings. Sci. Total Environ. 2015, 536, 964–972. [Google Scholar] [CrossRef]
- Delgado-Jarana, J.; Rincón, A.M.; Benı Tez, T.A. Aspartyl protease from Trichoderma harzianum CECT 2413: Cloning and characterization. Microbiology 2002, 148, 1305–1315. [Google Scholar] [CrossRef]
- Liaud, N.; Giniés, C.; Navarro, D.; Fabre, N.; Crapart, S.; Gimbert, I.H.; Levasseur, A.; Raouche, S.; Sigoillot, J.C. Exploring fungal biodiversity: Organic acid production by 66 strains of filamentous fungi. Fungal Biol. Biotechnol. 2014, 1, 1. [Google Scholar] [CrossRef]
- Dutton, M.; Evans, C. Oxalate production by fungi: Its role in pathogenicity and ecology in the soil environment. Can. J. Microbiol. 2011, 42, 881–895. [Google Scholar] [CrossRef]
- Plassard, C.; Fransson, P. Regulation of low-molecular weight organic acid production in fungi. Fungal Biol. Rev. 2009, 23, 30–39. [Google Scholar] [CrossRef]
- Hellström, P.; Heijnesson-Hultén, A.; Paulsson, M.; Håkansson, H.; Germgård, U. The effect of Fenton chemistry on the properties of microfibrillated cellulose. Cellulose 2014, 21, 1489–1503. [Google Scholar] [CrossRef]
- Shimada, M.; Akamtsu, Y.; Tokimatsu, T.; Mii, K.; Hattori, T. Possible biochemical roles of oxalic acid as a low molecular weight compound in-volved in brown-rot and white-rot wood decays. J. Biotechnol. 1997, 53, 103–113. [Google Scholar] [CrossRef]
- Viel, M.; Collet, F.; Lecieux, Y.; François, M.L.; Colson, V.; Lanos, C.; Hussain, A.; Lawrence, M. Resistance to mold development assessment of bio-based building materials. Compos. Part B Eng. 2019, 158, 406–418. [Google Scholar] [CrossRef]
- Construire en Chanvre. Règles professionnelles d’exécution; Collectif SEBTP; SEBTP: Paris, France, 2012; ISBN 978-2-35917-046-7. [Google Scholar]
- Tran, T.H.; Govin, A.; Guyonnet, R.; Grosseau, P.; Lors, C.; Damidot, D.; Deves, O.; Ruot, B. Influence of the intrinsic characteristics of mortars on their biofouling by pigmented organisms: Comparison between laboratory and field-scale experiments. Int. Biodeterior. Biodeterior. 2014, 86, 334–342. [Google Scholar] [CrossRef]
- Gangneux, C.; Akpa-Vinceslas, M.; Sauvage, H.; Desaire, S.; Houot, S.; Laval, K. Fungal, bacterial and plant dsDNA contributions to soil total DNA extracted from silty soils under different farming practices: Relationships with chloroform-labile carbon. Soil Biol. Biochem. 2011, 43, 431–437. [Google Scholar] [CrossRef]
- Borneman, J.; Hartin, R.J. PCR Primers That Amplify Fungal rRNA Genes from Environmental Samples. Appl. Environ. Microbiol. 2000, 66, 4356–4360. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Bartram, A.K.; Lynch, M.D.J.; Stearns, J.C.; Moreno-Hagelsieb, G.; Neufeld, J.D. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl. Environ. Microbiol. 2011, 77, 3846–3852. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R.; Bayesian, N. Clasifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, A.; Crous, P.W. Inside Plectosphaerellaceae. Stud. Mycol. 2019, 92, 227–286. [Google Scholar] [CrossRef] [PubMed]
- Grum-Grzhimaylo, A.A.; Georgieva, M.L.; Bondarenko, S.A.; Debets, A.J.M.; Bilanenko, E.N. On the diversity of fungi from soda soils. Fungal Divers. 2016, 76, 27–74. [Google Scholar] [CrossRef]
- Bandoni, R.J. The Tremellales and Auriculariales: An alternative classification. Trans. Mycol. Soc. Jpn. 1984, 25, 489–530. [Google Scholar]
- Diederich, P.; Ertz, D.; Eichler, M.; Cezanne, R.; Boom, P.; van den Broeck, D.V.; Serusiaux, E. New or interesting lichens and lichenicolous fungi from Belgium. Bull. Soc. Nat. Luxemb. 2014, 115, 157–165. [Google Scholar]
- Boekhout, T.; Fonseca, Á.; Sampaio, J.P.; Bandoni, R.J.; Fell, J.W.; Kwon-Chung, K.J. Chapter 100—Discussion of teleomorphic and anamorphic basidiomycetous yeasts. In Yeasts, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 1339–1372. [Google Scholar] [CrossRef]
- Liu, X.Z.; Wang, Q.M.; Theelen, B.; Groenewald, M.; Bai, F.Y.; Boekhout, T. Phylogeny of tremellomycetous yeasts and related dimorphic and fila-mentous basidiomycetes reconstructed from multiple gene sequence analyses. Stud. Mycol. 2015, 81, 1–26. [Google Scholar] [CrossRef]
- Mokhtarnejad, L.; Arzanlou, M.; Babai-Ahari, A.; Di Mauro, S.; Onofri, A.; Buzzini, P.; Turchetti, B. Characterization of basidiomycetous yeasts in hyper-saline soils of the Urmia Lake National Park, Iran. Extremophiles 2016, 20, 915–928. [Google Scholar] [CrossRef]
- Aust, H.J.; Bashi, E.; Rotem, J. Flexibility of plant pathogens in exploiting ecological and biotic conditions in the development of epidemics. In Comparative Epidemiology; Palti, J., Kranz, J., Eds.; A tool for better disease management; Pudoc: Wageningen, The Netherlands, 1980; pp. 46–56. [Google Scholar]
- Logrieco, A.; Moretti, A.; Solfrizzo, M. Alternaria toxins and plant diseases: An overview of origin, occurrence and risks. World Mycotoxin J. 2009, 2, 129–140. [Google Scholar] [CrossRef]
- Thomma, B.P. Alternaria spp.: From general saprophyte to specific parasite. Mol. Plant Pathol. 2003, 4, 225–236. [Google Scholar] [CrossRef]
- Lowy, B.A. Morphological Basis for Classifying the Species of Auricularia. Mycologia 1951, 43, 351–358. [Google Scholar] [CrossRef]
- Yuan, Y.; Wu, F.; Si, J.; Zhao, Y.F.; Dai, Y.C. Whole genome sequence of Auricularia heimuer (Basidiomycota, Fungi), the third most important cultivated mushroom worldwide. Genomics 2019, 111, 50–58. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kosiachevskyi, D.; Abahri, K.; Trinsoutrot-Gattin, I.; Castel, L.; Daubresse, A.; Chaouche, M.; Bennacer, R. New Refined Experimental Analysis of Fungal Growth in Degraded Bio-Based Materials. Processes 2024, 12, 2188. https://doi.org/10.3390/pr12102188
Kosiachevskyi D, Abahri K, Trinsoutrot-Gattin I, Castel L, Daubresse A, Chaouche M, Bennacer R. New Refined Experimental Analysis of Fungal Growth in Degraded Bio-Based Materials. Processes. 2024; 12(10):2188. https://doi.org/10.3390/pr12102188
Chicago/Turabian StyleKosiachevskyi, Dmytro, Kamilia Abahri, Isabelle Trinsoutrot-Gattin, Lisa Castel, Anne Daubresse, Mohend Chaouche, and Rachid Bennacer. 2024. "New Refined Experimental Analysis of Fungal Growth in Degraded Bio-Based Materials" Processes 12, no. 10: 2188. https://doi.org/10.3390/pr12102188
APA StyleKosiachevskyi, D., Abahri, K., Trinsoutrot-Gattin, I., Castel, L., Daubresse, A., Chaouche, M., & Bennacer, R. (2024). New Refined Experimental Analysis of Fungal Growth in Degraded Bio-Based Materials. Processes, 12(10), 2188. https://doi.org/10.3390/pr12102188







