Rapid and Non-Destructive Determination of Fatty Acid Profile and Oil Content in Diverse Brassica carinata Germplasm Using Fourier-Transform Near-Infrared Spectroscopy
Abstract
1. Introduction
2. Materials and Methods
2.1. Genetic Materials
2.2. Seed Sampling for NIRS and Reference Data
2.3. Spectra Data Acquisition and Analysis
2.4. Generation of Reference Data
2.4.1. Fatty Acid Profiling
2.4.2. Determination of Oil Content
2.5. Development of Calibration Model
3. Results and Discussion
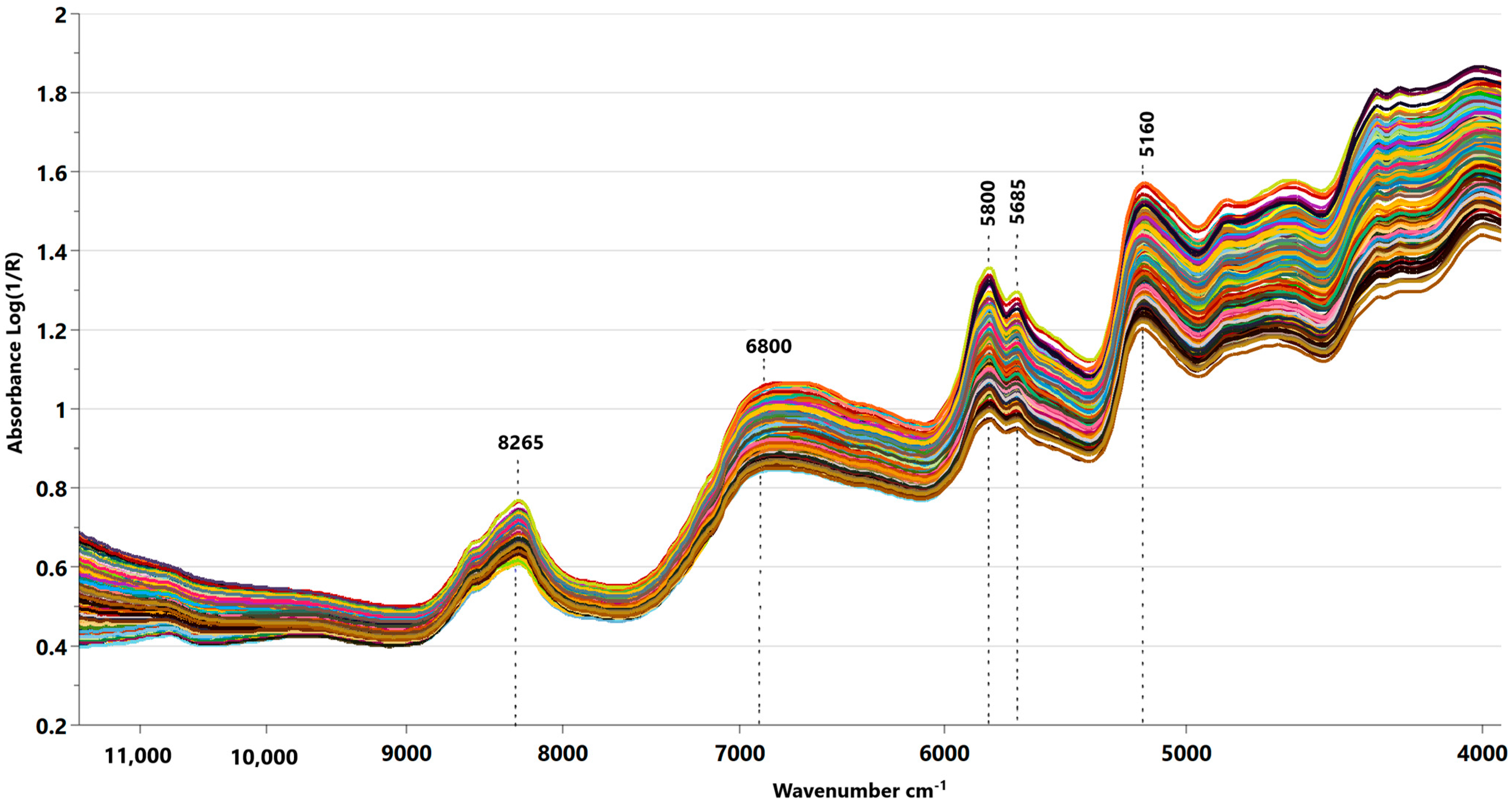
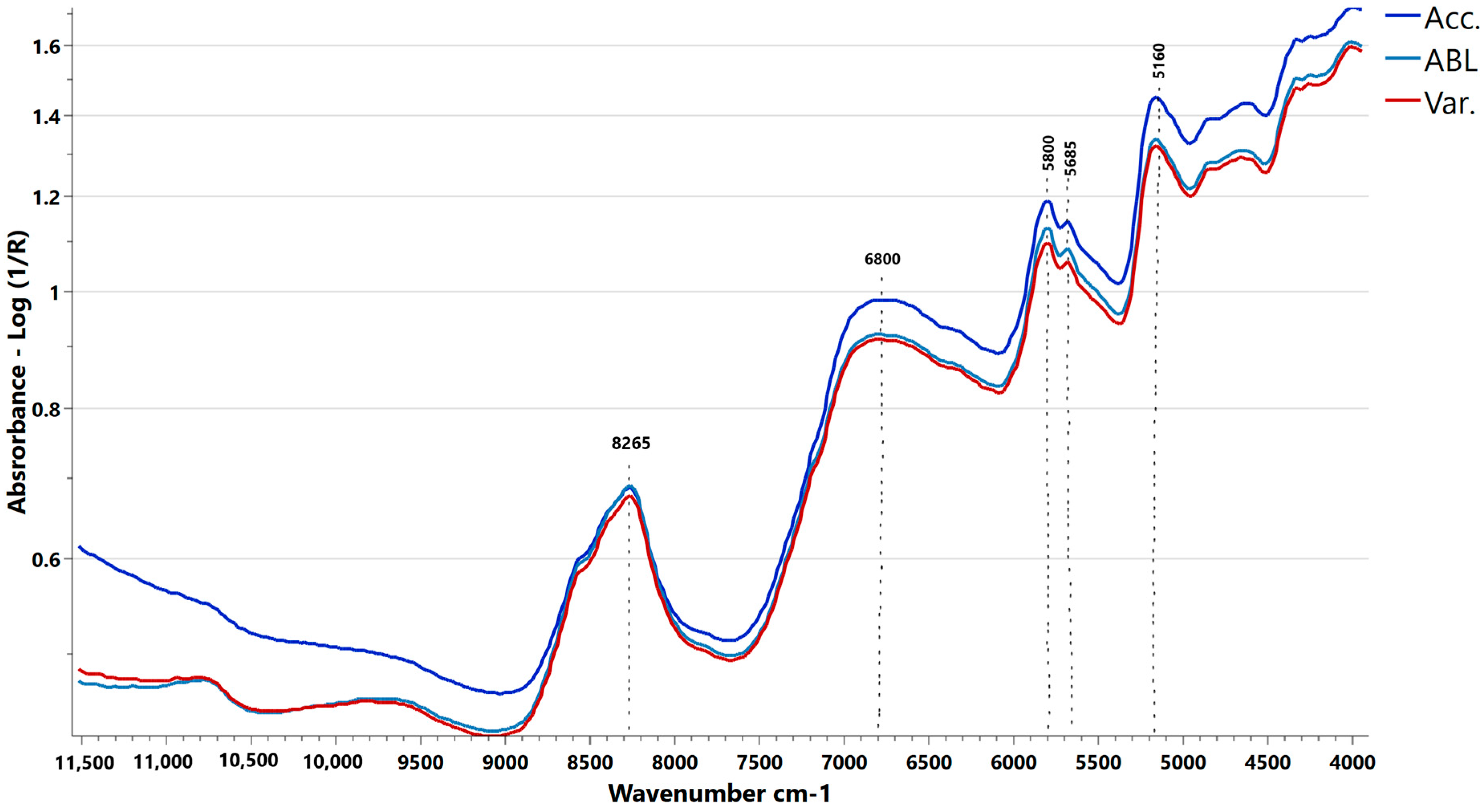
3.1. Spectra Data Analysis
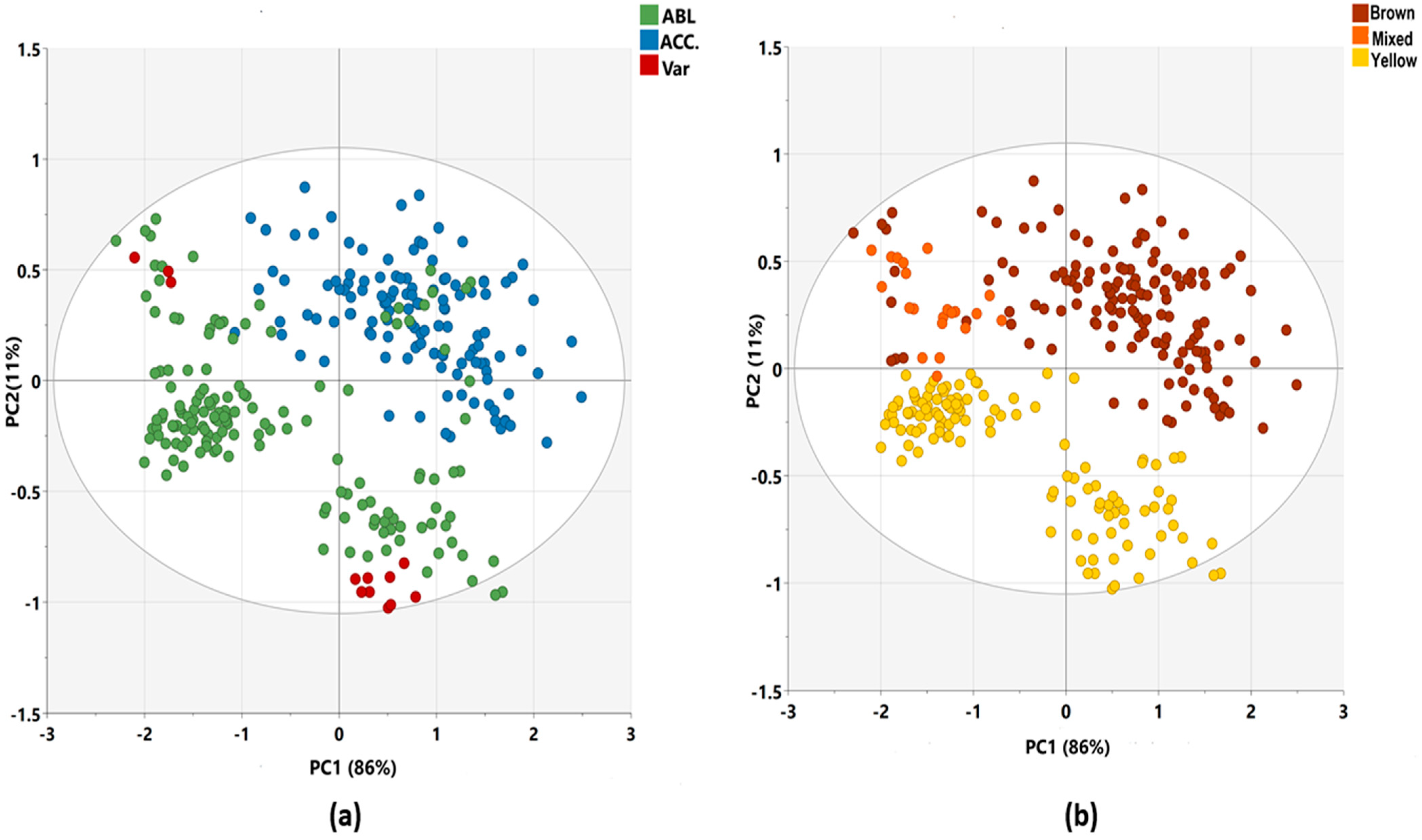
3.2. Principal Component Analysis (PCA)
3.3. Fatty Acid and Oil Content Analysis
3.4. Calibration Model Development
3.4.1. NIRS Spectra Pretreatment
3.4.2. NIRS Calibration Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taylor, D.C.; Falk, K.C.; Palmer, C.D.; Hammerlindl, J.; Babic, V.; Mietkiewska, E.; Jadhav, A.; Marillia, E.; Francis, T.; Hoffman, T.; et al. Brassica carinata–a new molecular farming platform for delivering bio-industrial oil feedstocks: Case studies of genetic modifications to improve very long-chain fatty acid and oil content in seeds. Biofuels Bioprod. Biorefining 2010, 4, 538–561. [Google Scholar] [CrossRef]
- Leonard, E.C. High-erucic vegetable oils. Ind. Crops Prod. 1992, 1, 119–123. [Google Scholar] [CrossRef]
- McVetty, P.B.; Scarth, R. Breeding for improved oil quality in Brassica oilseed species. J. Crop Prod. 2002, 5, 345–369. [Google Scholar] [CrossRef]
- Roslinsky, V.; Falk, K.C.; Gaebelein, R.; Mason, A.S.; Eynck, C. Development of B. carinata with super-high erucic acid content through interspecific hybridization. Theor. Appl. Genet. 2021, 134, 3167–3181. [Google Scholar] [CrossRef] [PubMed]
- Velasco, L.; Goffman, F.D.; Becker, H.C. Development of calibration equations to predict oil content and fatty acid composition in Brassicaceae germplasm by near-infrared reflectance spectroscopy. J. Am. Oil Chem. Soc. 1999, 76, 25–30. [Google Scholar] [CrossRef]
- Stefansson, B.; Hougen, F.W.; Downey, R. Note on the isolation of rape plants with seed oil free from erucic acid. Can. J. Plant Sci. 1961, 41, 218–219. [Google Scholar] [CrossRef]
- Getinet, A.; Rakow, G.; Raney, J.; Downey, R. Glucosinolate content in interspecific crosses of Brassica carinata with B. juncea and B. napus. Plant Breed. 1997, 116, 39–46. [Google Scholar] [CrossRef]
- Kumar, A.; Banga, S.S.; Meena, P.D.; Kumar, P.R. Brassica Oilseeds: Breeding and Management; CABI: Wallingford, UK, 2015. [Google Scholar]
- Ghanevati, M.; Jaworski, J.G. Engineering and mechanistic studies of the Arabidopsis FAE1 β-ketoacyl-CoA synthase, FAE1 KCS. Eur. J. Biochem. 2002, 269, 3531–3539. [Google Scholar] [CrossRef]
- Jadhav, A.; Katavic, V.; Marillia, E.F.; Giblin, E.M.; Barton, D.L.; Kumar, A.; Sonntag, C.; Babic, V.; Keller, W.A.; Taylor, D.C. Increased levels of erucic acid in Brassica carinata by co-suppression and antisense repression of the endogenous FAD2 gene. Metab. Eng. 2005, 7, 215–220. [Google Scholar] [CrossRef]
- Mietkiewska, E.; Giblin, E.M.; Wang, S.; Barton, D.L.; Dirpaul, J.; Brost, J.M.; Katavic, V.; Taylor, D.C. Seed-specific heterologous expression of a nasturtium FAE gene in Arabidopsis results in a dramatic increase in the proportion of erucic acid. Plant Physiol. 2004, 136, 2665–2675. [Google Scholar] [CrossRef][Green Version]
- Font, R.; del Río-Celestino, M.; de Haro-Bailón, A. The use of near-infrared spectroscopy (NIRS) in the study of seed quality components in plant breeding programs. Ind. Crops Prod. 2006, 24, 307–313. [Google Scholar] [CrossRef]
- Munck, L. A new holistic exploratory approach to systems biology by near infrared spectroscopy evaluated by chemometrics and data inspection. J. Chemom. A J. Chemom. Soc. 2007, 21, 406–426. [Google Scholar] [CrossRef]
- Montes, J.M.; Melchinger, A.E.; Reif, J.C. Novel throughput phenotyping platforms in plant genetic studies. Trends Plant Sci. 2007, 12, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Murray, I.; Cowe, I.A. Making Light Work: Advances in Near Infrared Spectroscopy; Wiley-VCH: Weinheim, Germany, 1992. [Google Scholar]
- Koester, S.; Paul, C. Application of Near Infrared Reflectance Spectroscopy (NIRS) in Breeding Rapeseed for Improved Quality; Vortraege fuer Pflanzenzuechtung: Göttingen, Germany, 1989. [Google Scholar]
- Oblath, E.A.; Isbell, T.A.; Berhow, M.A.; Allen, B.; Archer, D.; Brown, J.; Gesch, R.W.; Hatfield, J.L.; Jabro, J.D.; Kiniry, J.R.; et al. Development of near-infrared spectroscopy calibrations to measure quality characteristics in intact Brassicaceae germplasm. Ind. Crops Prod. 2016, 89, 52–58. [Google Scholar] [CrossRef][Green Version]
- Tkachuk, R. Oil and protein analysis of whole rapeseed kernels by near infrared reflectance spectroscopy. J. Am. Oil Chem. Soc. 1981, 58, 819–822. [Google Scholar] [CrossRef]
- Font, R.; Del Río, M.; Fernández, J.; Arthur, E.; Bancroft, I.; Chinoy, C.; De Haro, A. Seed oil content analysis of Ethiopian mustard (Brassica carinata A. Braun) by near infrared spectroscopy. Crucif. Newsl. 2002, 5–6. [Google Scholar]
- Petisco, C.; García-Criado, B.; Vázquez-De-Aldana, B.; de Haro, A.; García-Ciudad, A. Measurement of quality parameters in intact seeds of Brassica species using visible and near-infrared spectroscopy. Ind. Crops Prod. 2010, 32, 139–146. [Google Scholar] [CrossRef]
- Velasco, L.; Fernández-Martínez, J.; De Haro, A. Determination of the fatty acid composition of the oil in intact-seed mustard by near-infrared reflectance spectroscopy. J. Am. Oil Chem. Soc. 1997, 74, 1595–1602. [Google Scholar] [CrossRef]
- Pelc, S.E.; Couillard, D.M.; Stansell, Z.J.; Farnham, M.W. Genetic diversity and population structure of collard landraces and their relationship to other Brassica oleracea crops. Plant Genome 2015, 8, plantgenome2015-04. [Google Scholar] [CrossRef]
- Tesfaye, M.; AdugnaWakjira, A.T.; Terefe, G.; Weyessa, B.; Semahegn, Y. Achievements, Challenges and Future Prospects Edible Oilseeds Research and Development in Ethiopia. In Agricultural Research for Ethiopian Renaissance, Challenges, Opportunities and Directions; Proceedings of the National Conference on Agricultural Research for Ethiopian Renaissance held on January 26–27, 2016 in UNECA, Addis Ababa; Alemu, D., Derso, E., Asefa, G., Kirub, A., Eds.; Ethiopian Institute of Agricultuiral Research: Addis Ababa, Ethiopia, 2016; pp. 81–94. [Google Scholar]
- Deme, T.; Haki, G.D.; Retta, N.; Woldegiorgis, A.; Geleta, M. Fatty acid profile, total phenolic content, and antioxidant activity of niger seed (Guizotia abyssinica) and linseed (Linum usitatissimum). Front. Nutr. 2021, 8, 674882. [Google Scholar] [CrossRef]
- Williams, P.; Norris, K. Near-Infrared Technology in the Agricultural and Food Industries; American Association of Cereal Chemists, Inc.: St. Paul, MN, USA, 1987. [Google Scholar]
- Redda, Z.T.; Laß-Seyoum, A.; Yimam, A.; Barz, M.; Jabasingh, S.A. Solvent extraction and characterization of Brassica carinata oils as promising alternative feedstock for bio-jet fuel production. Biomass Convers. Biorefinery 2022, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Teklewold, A.; Becker, H.C. Geographic pattern of genetic diversity among 43 Ethiopian mustard (Brassica carinata A. Braun) accessions as revealed by RAPD analysis. Genet. Resour. Crop Evol. 2006, 53, 1173–1185. [Google Scholar] [CrossRef]
- Warwick, S.; Gugel, R.; McDonald, T.; Falk, K. Genetic variation of Ethiopian mustard (Brassica carinata A. Braun) germplasm in western Canada. Genet. Resour. Crop Evol. 2006, 53, 297–312. [Google Scholar] [CrossRef]
- Scarth, R.; Tang, J. Modification of Brassica oil using conventional and transgenic approaches. Crop Sci. 2006, 46, 1225–1236. [Google Scholar] [CrossRef]
- Seepaul, R.; Small, I.M.; Mulvaney, M.J.; George, S.; Leon, R.G.; Paula-Moraes, S.V.; Geller, D.; Marois, J.J.; Wright, D.L. Carinata, the Sustainable Crop for a Bio-Based Economy: 2018–2019 Production Recommendations for the Southeastern United States; University of Florida Institute of Food and Agricultural Sciences: Gainesville, FL, USA, 2019; pp. 1–12. [Google Scholar]
- Næs, T.; Isaksson, T.; Fearn, T.; Davies, T. A User-Friendly Guide to Multivariate Calibration and Classification; NIR: Chichester, UK, 2002. [Google Scholar]
- Williams, P.C.; Sobering, D. Comparison of commercial near infrared transmittance and reflectance instruments for analysis of whole grains and seeds. J. Near Infrared Spectrosc. 1993, 1, 25–32. [Google Scholar] [CrossRef]
- Williams, P. Implementation of near-infrared technology. In Near-Infrared Technology in the Agricultural and Food Industries, 2nd ed.; American Association of Cereal Chemists: St. Paul, MN, USA, 2001. [Google Scholar]
- Los, D.A.; Murata, N. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta BBA Lipids Lipid Metab. 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Taylor, D.C.; Francis, T.; Guo, Y.; Brost, J.M.; Katavic, V.; Mietkiewska, E.; Giblin, E.M.; Lozinsky, S.; Hoffman, T. Molecular cloning and characterization of a KCS gene from Cardamine graeca and its heterologous expression in Brassica oilseeds to engineer high nervonic acid oils for potential medical and industrial use. Plant Biotechnol. J. 2009, 7, 925–938. [Google Scholar] [CrossRef]
- Tillmann, P.; Reinhardt, T.-C.; Paul, C. Networking of near infrared spectroscopy instruments for rapeseed analysis: A comparison of different procedures. J. Near Infrared Spectrosc. 2000, 8, 101–107. [Google Scholar] [CrossRef]

| Varieties | Fatty Acid Profile (%) | Oil Content (%) | |||||
|---|---|---|---|---|---|---|---|
| C16:0 | C18:0 | C18:1 | C18:2 | C18:3 | C22:1 | ||
| Yellow Doddolla | 3.18 ± 0.10 b | 1.09 ± 0.07 a | 8.25 + 0.39 b | 19.78 + 0.15 a | 13.52 + 0.67 b | 42.79 + 0.23 a,b | 42.03 + 0.15 b |
| Dersash | 3.02 ± 0.03 b | 1.08 ± 0.03 a | 8.99 + 0.28 a | 19.88 + 0.01 a | 11.84 + 0.48 c | 41.74 + 0.13 b | 44.75 + 0.95 a |
| Tesfa | 3.01 ± 0.09 b | 1.09 ± 0.04 a | 8.25 + 0.29 b | 19.52 + 0.47 a | 12.58 + 0.34 b,c | 42.98 + 0.55 a | 41.62 + 0.73 b |
| Holetta-1 | 3.54 ± 0.09 a | 0.94 ± 0.02 b | 7.26 + 0.02 c | 18.60 + 0.25 b | 17.71 + 0.33 a | 39.17 + 0.62 c | 39.67 + 0.15 c |
| Fatty Acid or Oil Content | Range (%) | Mean (%) ± SD |
|---|---|---|
| C16:0 (palmitic acid) | 2.47–3.84 | 3.18 ± 0.25 |
| C18:0 (stearic acid) | 0.78–1.45 | 1.15 ± 0.11 |
| C18:1(oleic acid) | 7.23–12.60 | 9.22 ± 1.15 |
| C18:2 (linoleic acid) | 13.26–21.81 | 16.93 ± 1.41 |
| C18:3 (linolenic acid) | 9.68–22.65 | 14.64 ± 2.70 |
| C22:1 (erucic acid) | 34.93–48.10 | 41.66 ± 2.56 |
| OC (Oil content) | 26.89–48.23 | 36.88 ± 4.96 |
| Pretreatement | PCA1 (%) | PCA2 (%) | Number of Outlier Samples |
|---|---|---|---|
| SNV + SG smoothing | 76.1 | 4.2 | 4 |
| MSC + SG smoothing | 76.1 | 4.1 | 5 |
| SNV + SGD1 | 52.3 | 27.2 | 0 |
| MSC + SGD2 | 51.5 | 25.4 | 1 |
| Fatty Acid and Oil Content | Calibration Staistics | Validation Statistics | |||
|---|---|---|---|---|---|
| R2cal | RMSEC | R2val | RMSEP | RPD | |
| C16:0 (palmitic acid) | 0.64 | 0.13 | 0.78 | 0.12 | 2.1 |
| C18:0 (stearic acid) | 0.67 | 0.05 | 0.75 | 0.06 | 2.0 |
| C18:1(oleic acid) | 0.88 | 0.39 | 0.92 | 0.22 | 3.6 |
| C18:2 (linoleic acid) | 0.90 | 0.45 | 0.89 | 0.27 | 3.2 |
| C18:3 (linolenic acid) | 0.86 | 0.89 | 0.93 | 0.59 | 3.8 |
| C22:1 (erucic acid) | 0.87 | 0.89 | 0.92 | 0.57 | 3.5 |
| OC (oil content) | 0.93 | 0.82 | 0.93 | 1.41 | 3.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tesfaye, M.; Feyissa, T.; Hailesilassie, T.; Wang, E.S.; Kanagarajan, S.; Zhu, L.-H. Rapid and Non-Destructive Determination of Fatty Acid Profile and Oil Content in Diverse Brassica carinata Germplasm Using Fourier-Transform Near-Infrared Spectroscopy. Processes 2024, 12, 244. https://doi.org/10.3390/pr12020244
Tesfaye M, Feyissa T, Hailesilassie T, Wang ES, Kanagarajan S, Zhu L-H. Rapid and Non-Destructive Determination of Fatty Acid Profile and Oil Content in Diverse Brassica carinata Germplasm Using Fourier-Transform Near-Infrared Spectroscopy. Processes. 2024; 12(2):244. https://doi.org/10.3390/pr12020244
Chicago/Turabian StyleTesfaye, Misteru, Tileye Feyissa, Teklehaimanot Hailesilassie, Eu Sheng Wang, Selvaraju Kanagarajan, and Li-Hua Zhu. 2024. "Rapid and Non-Destructive Determination of Fatty Acid Profile and Oil Content in Diverse Brassica carinata Germplasm Using Fourier-Transform Near-Infrared Spectroscopy" Processes 12, no. 2: 244. https://doi.org/10.3390/pr12020244
APA StyleTesfaye, M., Feyissa, T., Hailesilassie, T., Wang, E. S., Kanagarajan, S., & Zhu, L.-H. (2024). Rapid and Non-Destructive Determination of Fatty Acid Profile and Oil Content in Diverse Brassica carinata Germplasm Using Fourier-Transform Near-Infrared Spectroscopy. Processes, 12(2), 244. https://doi.org/10.3390/pr12020244






