MYB Transcriptional Factors Affects Upstream and Downstream MEP Pathway and Triterpenoid Biosynthesis in Chlamydomonas reinhardtii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain of C. reinhardtii and Culture Condition
2.2. The Construction of Plasmid and Expression Screening
2.3. Transformation and Screening
2.4. Genomic DNA and RNA Extraction and cDNA Synthesis
2.5. The Analysis of qPCR and Genomic PCR
2.6. Measurements of Squalene Contents
2.7. Statistical Analysis
3. Results
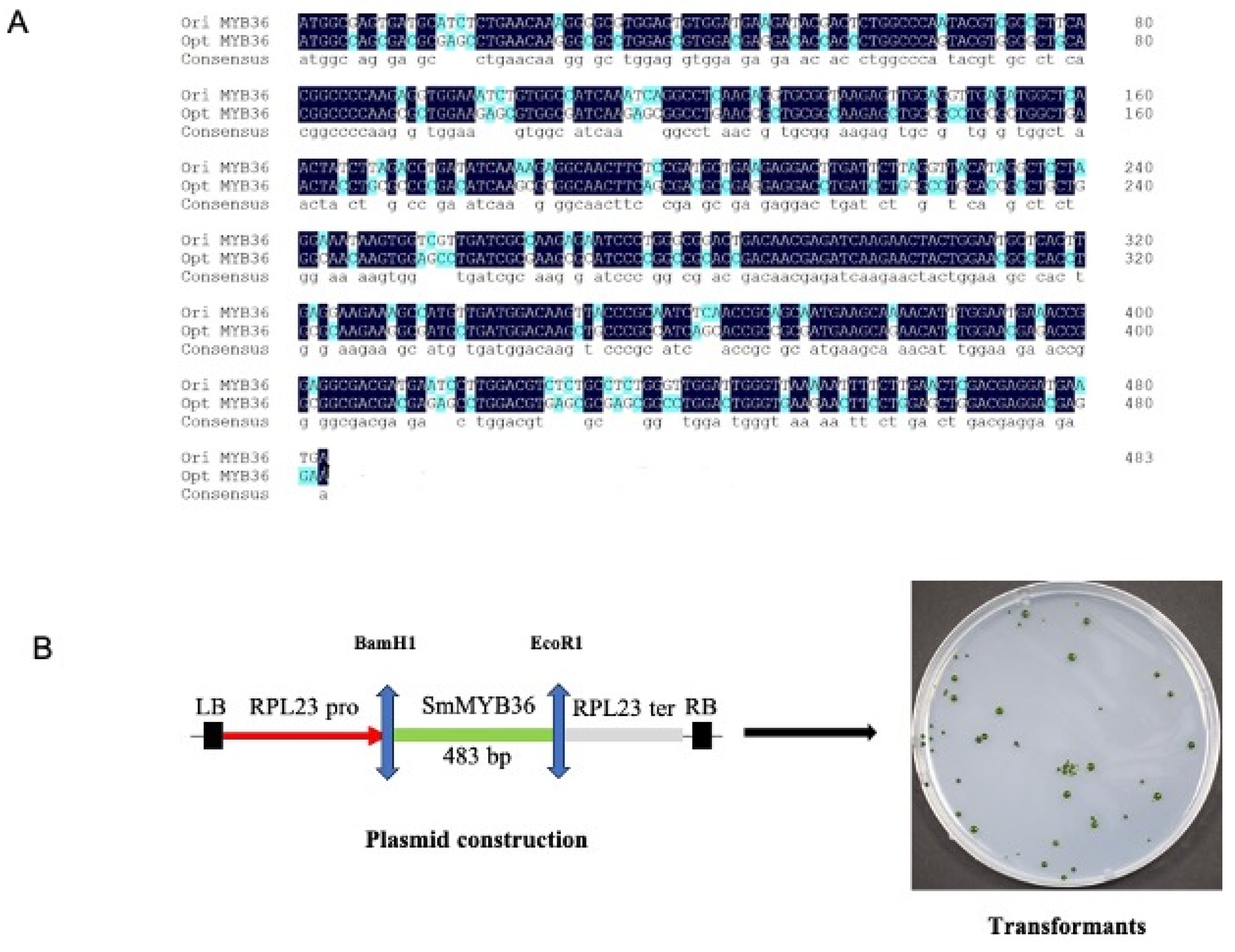
3.1. The Design Expression Cassette of MYB36
3.2. The Screening of Transgenic Strain of C. reinhardtii
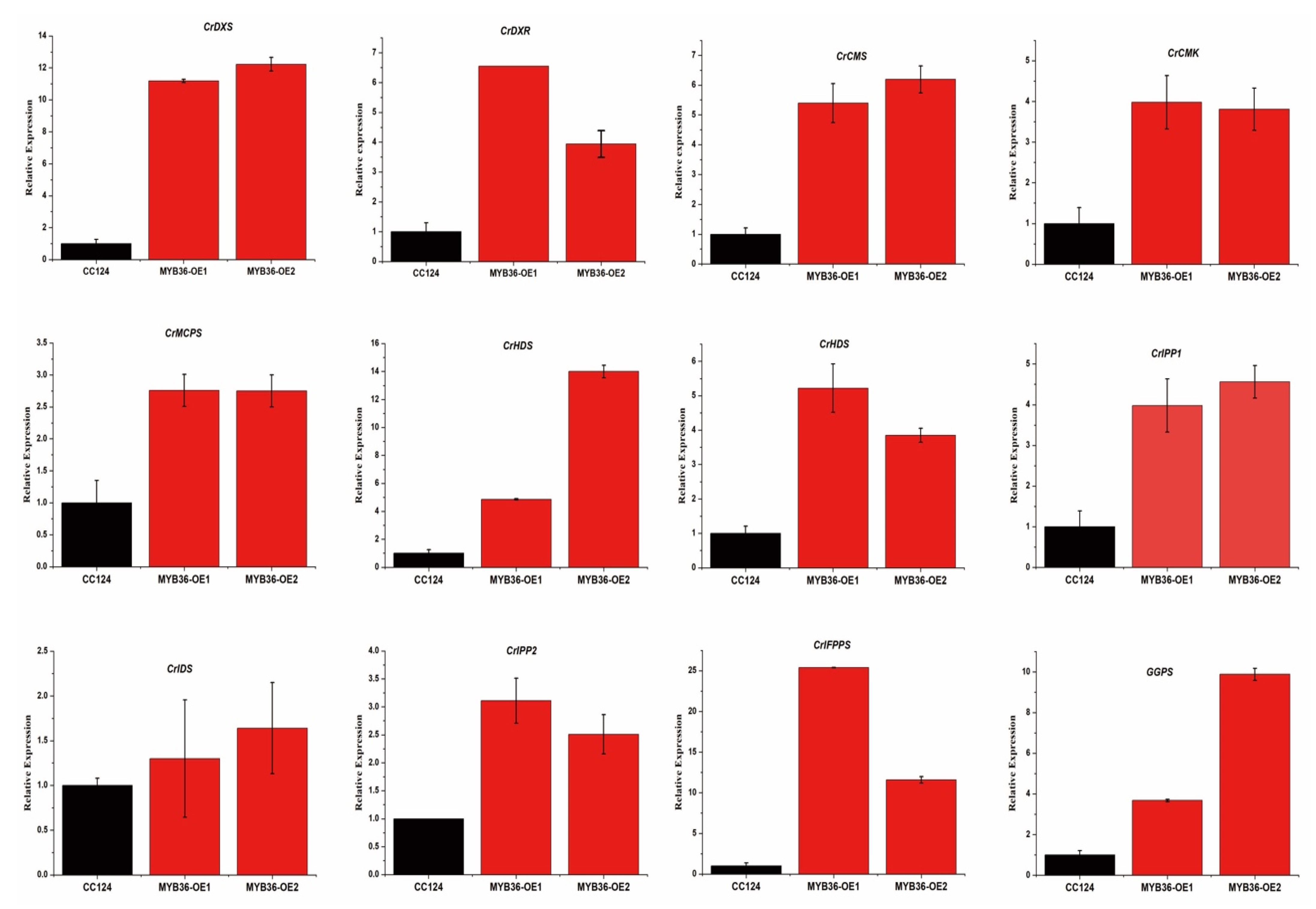
3.3. MYB36 Transcriptional Factors Upregulate the MEP Biosynthetic Pathway in Overexpression Lines of C. reinhardtii
3.4. MYB 36 Affect the Endogenous Squalene Biosynthetic Pathway Genes
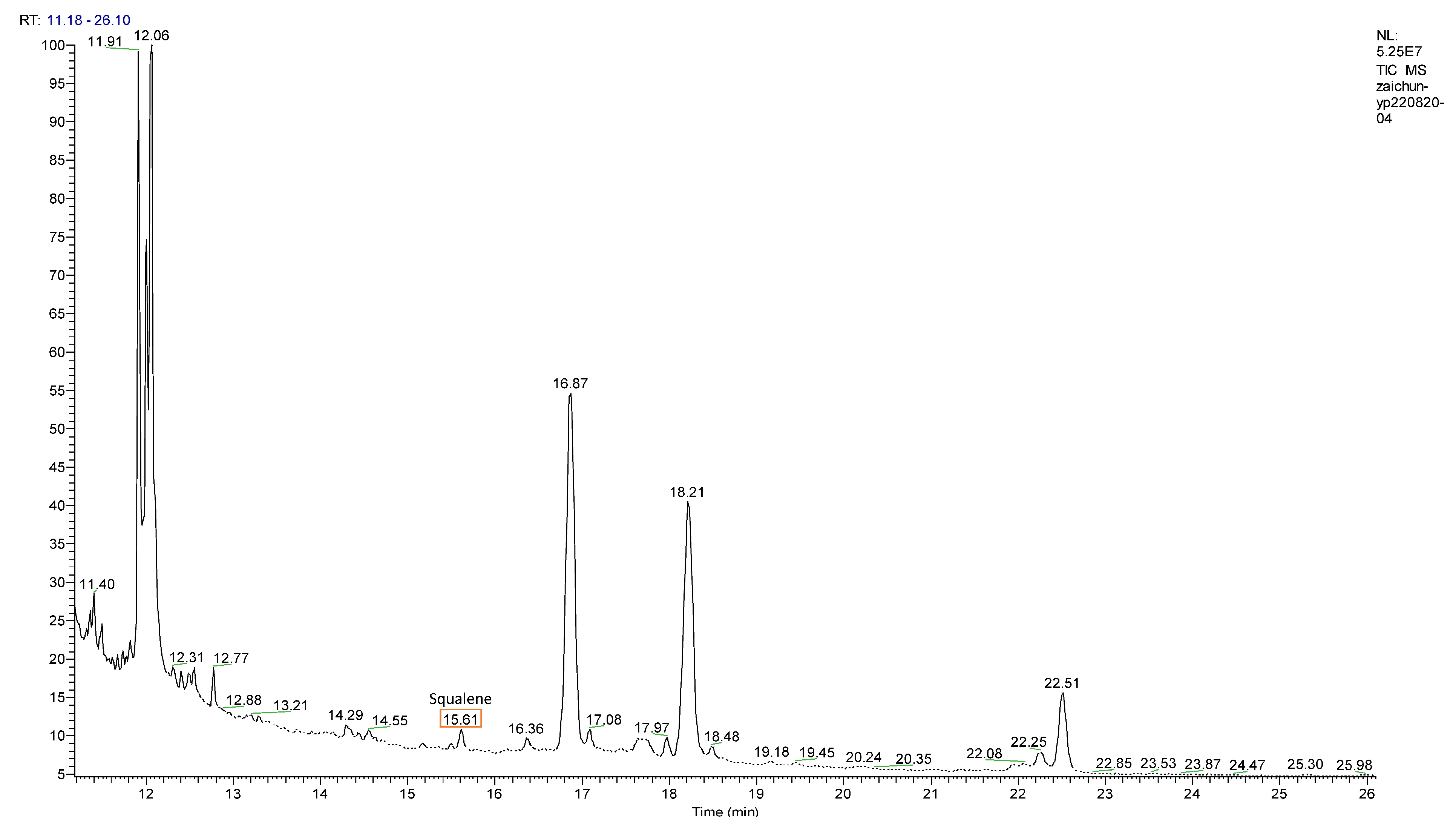
3.5. Accumulation of Squalene on MYB-Overexpression Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Onyeaka, H.; Miri, T.; Obileke, K.; Hart, A.; Anumudu, C.; Al-Sharify, Z.T. Minimizing carbon footprint via microalgae as a biological capture. Carbon Capture Sci. Technol. 2021, 1, 100007. [Google Scholar] [CrossRef]
- Blas-Valdivia, V.; Ortiz-Butrón, R.; Pineda-Reynoso, M.; Hernández-Garcia, A.; Cano-Europa, E. Chlorella vulgaris administration prevents HgCl 2-caused oxidative stress and cellular damage in the kidney. J. Appl. Phycol. 2011, 23, 53–58. [Google Scholar] [CrossRef]
- Torres-Tiji, Y.; Fields, F.J.; Mayfield, S.P. Microalgae as a future food source. Biotechnol. Adv. 2020, 41, 107536. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Saha, P.; Rai, N.; Kumari, S.; Pandey-Rai, S. Unravelling triterpenoid biosynthesis in plants for applications in bioengineering and large-scale sustainable production. Ind. Crops Prod. 2023, 199, 116789. [Google Scholar] [CrossRef]
- Vickers, C.E.; Williams, T.C.; Peng, B.; Cherry, J. Recent advances in synthetic biology for engineering isoprenoid production in yeast. Curr. Opin. Chem. Biol. 2017, 40, 47–56. [Google Scholar] [CrossRef]
- Vickers, C.E.; Behrendorff, J.B.; Bongers, M.; Brennan, T.C.; Bruschi, M.; Nielsen, L.K. Production of industrially relevant isoprenoid compounds in engineered microbes. In Microorganisms in Biorefineries; Springer: Berlin/Heidelberg, Germany, 2015; pp. 303–334. [Google Scholar]
- Masi, A.; Leonelli, F.; Scognamiglio, V.; Gasperuzzo, G.; Antonacci, A.; Terzidis, M.A. Chlamydomonas reinhardtii: A factory of nutraceutical and food supplements for human health. Molecules 2023, 28, 1185. [Google Scholar] [CrossRef] [PubMed]
- Yahya, R.Z.; Wellman, G.B.; Overmans, S.; Lauersen, K.J. Engineered production of isoprene from the model green microalga Chlamydomonas reinhardtii. Metab. Eng. Commun. 2023, 16, e00221. [Google Scholar] [CrossRef]
- Sun, J.; Xu, X.; Wu, Y.; Sun, H.; Luan, G.; Lu, X. Conversion of carbon dioxide into valencene and other sesquiterpenes with metabolic engineered Synechocystis sp. PCC 6803 cell factories. GCB Bioenergy 2023, 15, 1154–1165. [Google Scholar] [CrossRef]
- Xie, Z.; He, J.; Peng, S.; Zhang, X.; Kong, W. Biosynthesis of protein-based drugs using eukaryotic microalgae. Algal Res. 2023, 74, 103219. [Google Scholar] [CrossRef]
- Spanova, M.; Daum, G. Squalene–biochemistry, molecular biology, process biotechnology, and applications. Eur. J. Lipid Sci. Technol. 2011, 113, 1299–1320. [Google Scholar] [CrossRef]
- Ibrahim, N.I.; Fairus, S.; Zulfarina, M.S.; Naina Mohamed, I. The efficacy of squalene in cardiovascular disease risk-a systematic review. Nutrients 2020, 12, 414. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Karadeniz, F. Biological importance and applications of squalene and squalane. Adv. Food Nutr. Res. 2012, 65, 223–233. [Google Scholar]
- Pacetti, D.; Scortichini, S.; Boarelli, M.C.; Fiorini, D. Simple and rapid method to analyse squalene in olive oils and extra virgin olive oils. Food Control 2019, 102, 240–244. [Google Scholar] [CrossRef]
- Nocito, F.; Labrador Garcia, A. Towards New Sustainable Squalene Resources: Extraction from Apulian “Aged Extra-Virgin Olive Oil Sludge”(AEVOO-S). A Comparison Between Organic Solvent and Supercritical Fluid Techniques. Waste Biomass Valorization 2023, 14, 2275–2284. [Google Scholar] [CrossRef]
- Acosta-Martínez, V.; Burow, G.; Zobeck, T.; Allen, V. Soil microbial communities and function in alternative systems to continuous cotton. Soil Sci. Soc. Am. J. 2010, 74, 1181–1192. [Google Scholar] [CrossRef]
- Waterman, E.; Lockwood, B. Active components and clinical applications of olive oil. Altern. Med. Rev. 2007, 12, 331–342. [Google Scholar]
- Song, X.; Wang, X.; Tan, Y.; Feng, Y.; Li, W.; Cui, Q. High production of squalene using a newly isolated yeast-like strain Pseudozyma sp. SD301. J. Agric. Food Chem. 2015, 63, 8445–8451. [Google Scholar] [CrossRef]
- Paramasivan, K.; Mutturi, S. Recent advances in the microbial production of squalene. World J. Microbiol. Biotechnol. 2022, 38, 91. [Google Scholar] [CrossRef]
- Bhattacharjee, P.; Shukla, V.; Singhal, R.; Kulkarni, P. Studies on fermentative production of squalene. World J. Microbiol. Biotechnol. 2001, 17, 811–816. [Google Scholar] [CrossRef]
- Nakazawa, A.; Matsuura, H.; Kose, R.; Kato, S.; Honda, D.; Inouye, I.; Kaya, K.; Watanabe, M.M. Optimization of culture conditions of the thraustochytrid Aurantiochytrium sp. strain 18W-13a for squalene production. Bioresour. Technol. 2012, 109, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.K.; Heo, S.Y.; Park, H.M.; Kim, C.H.; Sohn, J.H.; Kondo, A.; Seo, J.W. Characterization of a squalene synthase from the thraustochytrid microalga Aurantiochytrium sp. krs101. J. Microbiol. Biotechnol. 2013, 23, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Lohr, M.; Schwender, J.; Polle, J.E. Isoprenoid biosynthesis in eukaryotic phototrophs: A spotlight on algae. Plant Sci. 2012, 185–186, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Lauersen, K.J.; Wichmann, J.; Baier, T.; Kampranis, S.C.; Pateraki, I.; Møller, B.L.; Kruse, O. Phototrophic production of heterologous diterpenoids and a hydroxy-functionalized derivative from Chlamydomonas reinhardtii. Metab. Eng. 2018, 49, 116–127. [Google Scholar] [CrossRef]
- Davies, F.K.; Jinkerson, R.E.; Posewitz, M.C. Toward a photosynthetic microbial platform for terpenoid engineering. Photosynth. Res. 2015, 123, 265–284. [Google Scholar] [CrossRef] [PubMed]
- Mahjoub, A.; Hernould, M.; Joubès, J.; Decendit, A.; Mars, M.; Barrieu, F.; Hamdi, S.; Delrot, S. Overexpression of a grapevine R2R3-MYB factor in tomato affects vegetative development, flower morphology and flavonoid and terpenoid metabolism. Plant Physiol. Biochem. 2009, 47, 551–561. [Google Scholar] [CrossRef]
- Li, L.; Wang, D.; Zhou, L.; Yu, X.; Yan, X.; Zhang, Q.; Li, B.; Liu, Y.; Zhou, W.; Cao, X.; et al. JA-responsive transcription factor SmMYB97 promotes phenolic acid and tanshinone accumulation in Salvia miltiorrhiza. J. Agric. Food Chem. 2020, 68, 14850–14862. [Google Scholar] [CrossRef]
- Hao, X.; Pu, Z.; Cao, G.; You, D.; Zhou, Y.; Deng, C.; Shi, M.; Nile, S.H.; Wang, Y.; Zhou, W.; et al. Tanshinone and salvianolic acid biosynthesis are regulated by SmMYB98 in Salvia miltiorrhiza hairy roots. J. Adv. Res. 2020, 23, 1–12. [Google Scholar] [CrossRef]
- He, Y.; Li, M.; Wang, Y.; Shen, S. The R2R3-MYB transcription factor MYB44 modulates carotenoid biosynthesis in Ulva prolifera. Algal Res. 2022, 62, 102578. [Google Scholar] [CrossRef]
- Kong, F.; Yamasaki, T.; Ohama, T. Expression levels of domestic cDNA cassettes integrated in the nuclear genomes of various Chlamydomonas reinhardtii strains. J. Biosci. Bioeng. 2014, 117, 613–616. [Google Scholar] [CrossRef]
- Pandit, J.; Danley, D.E.; Schulte, G.K.; Mazzalupo, S.; Pauly, T.A.; Hayward, C.M.; Hamanaka, E.S.; Thompson, J.F.; Harwood, H.J. Crystal structure of human squalene synthase: A key enzyme in cholesterol biosynthesis. J. Biol. Chem. 2000, 275, 30610–30617. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Huang, R.; Wang, C.; Hu, Q.; Li, H.; Li, X. Expression of anti-lipopolysaccharide factor isoform 3 in Chlamydomonas reinhardtii showing high antimicrobial activity. Mar. Drugs 2021, 19, 239. [Google Scholar] [CrossRef] [PubMed]
- Lange, B.M.; Rujan, T.; Martin, W.; Croteau, R. Isoprenoid biosynthesis: The evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. USA 2000, 97, 13172–13177. [Google Scholar] [CrossRef] [PubMed]
- Vranová, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef]
- Estévez, J.M.; Cantero, A.; Reindl, A.; Reichler, S.; León, P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 2001, 276, 22901–22909. [Google Scholar] [CrossRef] [PubMed]
- Cordoba, E.; Salmi, M.; León, P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 2009, 60, 2933–2943. [Google Scholar] [CrossRef] [PubMed]
- Commault, A.S.; Fabris, M.; Kuzhiumparambil, U.; Adriaans, J.; Pernice, M.; Ralph, P.J. Methyl jasmonate treatment affects the regulation of the 2-C-methyl-D-erythritol 4-phosphate pathway and early steps of the triterpenoid biosynthesis in Chlamydomonas reinhardtii. Algal Res. 2019, 39, 101462. [Google Scholar] [CrossRef]
- Banerjee, A.; Sharkey, T. Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat. Prod. Rep. 2014, 31, 1043–1055. [Google Scholar] [CrossRef]
- Brumfield, K.M.; Laborde, S.M.; Moroney, J.V. A model for the ergosterol biosynthetic pathway in Chlamydomonas reinhardtii. Eur. J. Phycol. 2017, 52, 64–74. [Google Scholar] [CrossRef]
- Kajikawa, M.; Kinohira, S.; Ando, A.; Shimoyama, M.; Kato, M.; Fukuzawa, H. Accumulation of squalene in a microalga Chlamydomonas reinhardtii by genetic modification of squalene synthase and squalene epoxidase genes. PLoS ONE 2015, 10, e0120446. [Google Scholar] [CrossRef]
- Anwar, M.; Duan, S.; Ma, M.; Chen, X.; Wu, L.; Zeng, L. NataMYB4, a flower specific gene, regulates the flavonoid biosynthesis in Chinese Narcissus. Sci. Hortic. 2023, 318, 112101. [Google Scholar] [CrossRef]
- Anwar, M.; Chen, L.; Xiao, Y.; Wu, J.; Zeng, L.; Li, H.; Wu, Q.; Hu, Z. Recent advanced metabolic and genetic engineering of phenylpropanoid biosynthetic pathways. Int. J. Mol. Sci. 2021, 22, 9544. [Google Scholar] [CrossRef]
- Yin, J.; Li, X.; Zhan, Y.; Li, Y.; Qu, Z.; Sun, L.; Wang, S.; Yang, J.; Xiao, J. Cloning and expression of BpMYC4 and BpbHLH9 genes and the role of BpbHLH9 in triterpenoid synthesis in birch. BMC Plant Biol. 2017, 17, 214. [Google Scholar] [CrossRef]
- Wei, J.; Yang, Y.; Peng, Y.; Wang, S.; Zhang, J.; Liu, X.; Liu, J.; Wen, B.; Li, M. Biosynthesis and the Transcriptional Regulation of Terpenoids in Tea Plants (Camellia sinensis). Int. J. Mol. Sci. 2023, 24, 6937. [Google Scholar] [CrossRef]
- Thiriet-Rupert, S.; Carrier, G.; Trottier, C.; Eveillard, D.; Schoefs, B.; Bougaran, G.; Cadoret, J.-P.; Chénais, B.; Saint-Jean, B. Identification of transcription factors involved in the phenotype of a domesticated oleaginous microalgae strain of Tisochrysis lutea. Algal Res. 2018, 30, 59–72. [Google Scholar] [CrossRef]
- Xia, M.; Tu, L.; Liu, Y.; Jiang, Z.; Wu, X.; Gao, W.; Huang, L. Genome-wide analysis of MYB family genes in Tripterygium wilfordii and their potential roles in terpenoid biosynthesis. Plant Direct 2022, 6, e424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, X.; Yu, X.; Wang, F.; Long, J.; Shen, W.; Jiang, D.; Zhao, X. The MYB transcription factor CiMYB42 regulates limonoids biosynthesis in citrus. BMC Plant Biol. 2020, 20, 254. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Pei, T.; Bai, Z.; Jia, Y.; Ma, P.; Liang, Z. SmMYB36, a novel R2R3-MYB transcription factor, enhances tanshinone accumulation and decreases phenolic acid content in Salvia miltiorrhiza hairy roots. Sci. Rep. 2017, 7, 5104. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Liu, G.; Yang, M.; Wang, X.; Chen, X.; Chen, F.; Yang, Y. Isolation and functional analysis of squalene synthase gene in tea plant Camellia sinensis. Plant Physiol. Biochem. 2019, 142, 53–58. [Google Scholar] [CrossRef] [PubMed]


| Genes | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| CrDXS | GACGGTGGCTATGCACTATG | GAAATCGAGGTGGAGCTGTG |
| CrDXR | CCATCTTCCAGGTGATGCAG | CCAGACCCTTGTTCATGAGTG |
| CrCMS | GCACTACTGCTCCTACCAAG | GAACGTCTCCAGCGAGTATG |
| CrCMK | ACGCTGCAGACCATGTACTA | GTTGGAGAAGAAGACCGAGATG |
| CrMECPS | CTCTGTGCCTCCCAGACATC | GTTGCGGATGTTCTCCTTGTG |
| CHDS | GCTGATTGAGGAGACCTTAC | GCAGAAGACGAAGTTGTGGTAG |
| CrHDR | CTGACTGACTTCAAGGAGAAGG | GATGTAGTTGGAGGCGAAGG |
| CrIPPI | TCCTCCTTCTCCTTCCTCAC | GCCATCATGGACTGAAGCTC |
| CrIPP2 | TTCCGCAACAAGGGATTCAG | GAACCAGACCGTGTAGTCCT |
| CrGPS | GTGCTGTCGCTCAATACCAG | AGTAGGTCTTGGCCAGGTAG |
| CrFPS | CCGAGGATGAGGTGTTCAAG | CGCTTGAGGATGGAGTAGATG |
| CrGGPS | CCATGGACAACGACGACTTC | TTGCCCAGCTCCATGATTAC |
| CrSQS | TGGCAGCATGCTACAACAAC | GAACTGCAGGAACCAGGTGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anwar, M.; Wang, J.; Li, J.; Altaf, M.M.; Hu, Z. MYB Transcriptional Factors Affects Upstream and Downstream MEP Pathway and Triterpenoid Biosynthesis in Chlamydomonas reinhardtii. Processes 2024, 12, 487. https://doi.org/10.3390/pr12030487
Anwar M, Wang J, Li J, Altaf MM, Hu Z. MYB Transcriptional Factors Affects Upstream and Downstream MEP Pathway and Triterpenoid Biosynthesis in Chlamydomonas reinhardtii. Processes. 2024; 12(3):487. https://doi.org/10.3390/pr12030487
Chicago/Turabian StyleAnwar, Muhammad, Jingkai Wang, Jiancheng Li, Muhammad Mohsin Altaf, and Zhangli Hu. 2024. "MYB Transcriptional Factors Affects Upstream and Downstream MEP Pathway and Triterpenoid Biosynthesis in Chlamydomonas reinhardtii" Processes 12, no. 3: 487. https://doi.org/10.3390/pr12030487
APA StyleAnwar, M., Wang, J., Li, J., Altaf, M. M., & Hu, Z. (2024). MYB Transcriptional Factors Affects Upstream and Downstream MEP Pathway and Triterpenoid Biosynthesis in Chlamydomonas reinhardtii. Processes, 12(3), 487. https://doi.org/10.3390/pr12030487







