Response Surface Methodology Applied to Cyanobacterial EPS Production: Steps and Statistical Validations
Abstract
1. Introduction
- Step 1—Experimental design—creation of fractional factorial production design;
- Step 2—Polynomial model—fitting of collected data into a second-order polynomial model;
- Step 3—Model validation—a statistical evaluation of the model’s validity and the assessment of the significance of the involved independent variables;
- Step 4—Response optimization—identification of optimal independent variables to secure the best possible outcomes.
2. Experimental Design
2.1. S-EPS Production and Determination
2.2. RSM Methodology
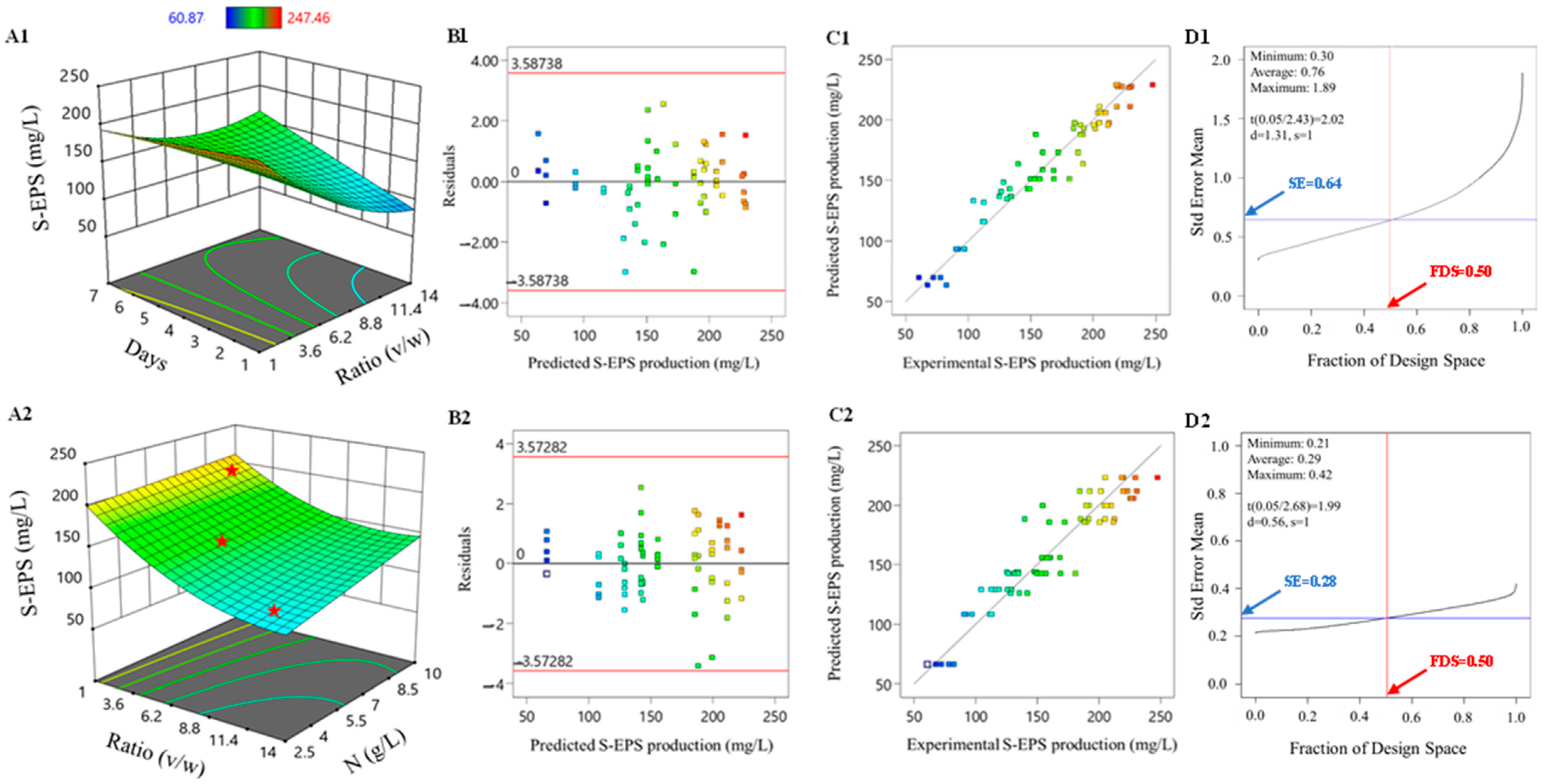
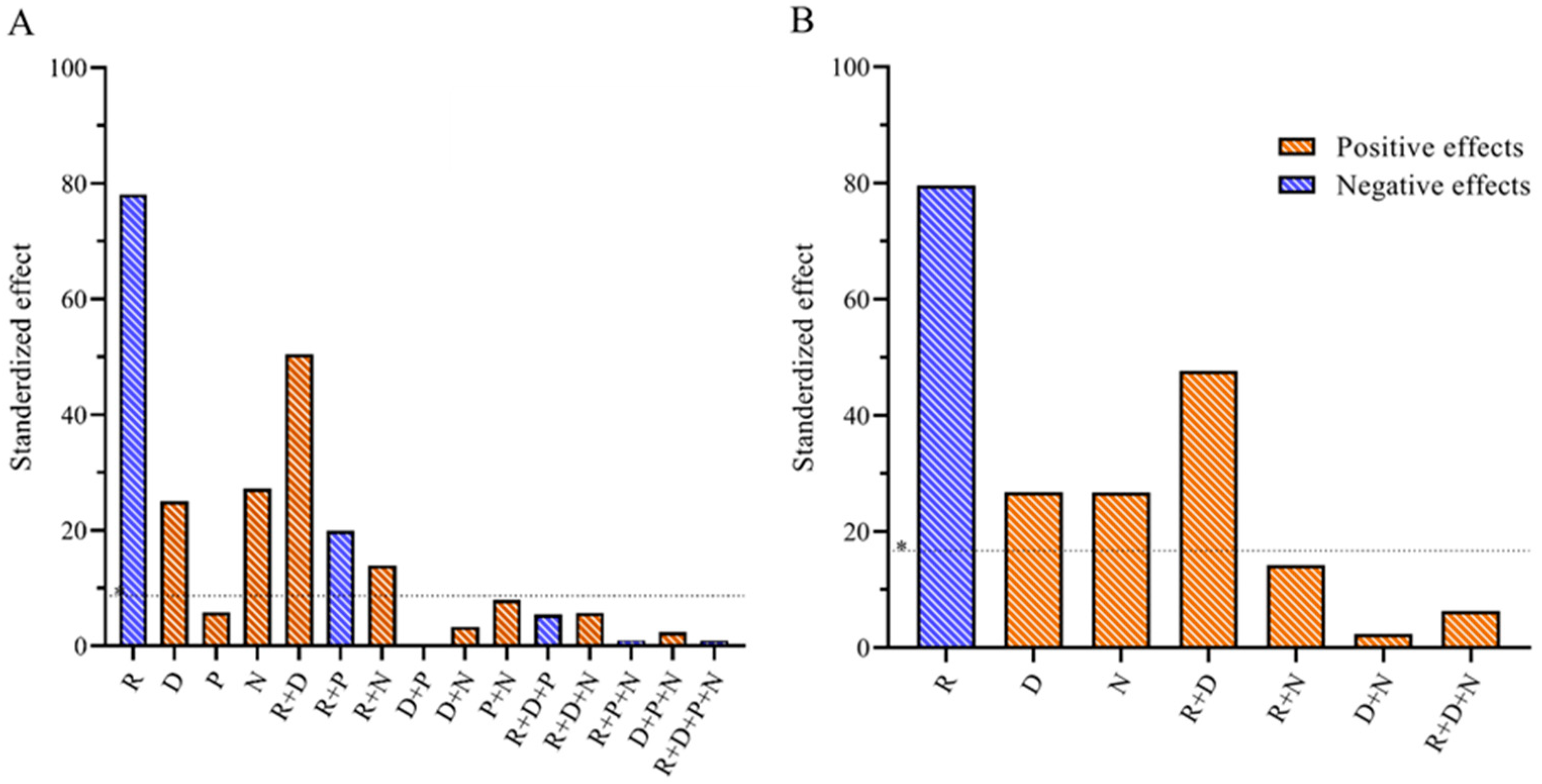
3. Results and Discussion
3.1. RSM Model: Application, Refinement, and Validation Applications to Response Optimization
3.2. RSM Model: Applications to Response Optimization
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cruz, D.; Vasconcelos, V.; Pierre, G.; Michaud, P.; Delattre, C. Exopolysaccharides from cyanobacteria: Strategies for bioprocess development. Appl. Sci. 2020, 10, 3763. [Google Scholar] [CrossRef]
- Tiwari, O.N.; Khangembam, R.; Shamjetshabam, M.; Sharma, A.S.; Oinam, G.; Brand, J.J. Characterisation and optimisation of bioflocculant exopolysaccharide production by cyanobacteria Nostoc sp. BTA97 and Anabaena sp. BTA990 in culture conditions. Appl. Biochem. Biotechnol. 2015, 176, 1950–1963. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Shera, S.S.; Banik, J.; Banik, R.M. Optimisation of cultural conditions using response surface methodology versus artificial neural network and modelling of L-glutaminase production by Bacillus cereus MTCC 1305. Bioresour. Technol. 2013, 137, 261–269. [Google Scholar] [CrossRef]
- Reig, S.J.M.; Luti, K.J.K. Response surface methodology: A review on its applications and challenges in microbial cultures. Mater. Today Proc. 2021, 42, 2277–2284. [Google Scholar] [CrossRef]
- Domagalski, N.R.; Mack, B.C.; Tabora, J.E. Analysis of design of experiments with dynamic responses. Org. Process Res. Dev. 2015, 19, 1667–1682. [Google Scholar] [CrossRef]
- Meyers, R.H.; Montgomery, D.C.; Anderson-Cook, M.A. Response surface methodology: Process and product optimisation using designed experiments. In Response Surface Methodology; Balding, D.J., Cressie, N.A.C., Fitzmaurice, G.M., Johnstone, I.M., Molenberghs, G., Scott, D.W., Smith, A.F.M., Tsay, R.S., Weisberg, S., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009. [Google Scholar]
- Rodrigues, F.; Faria, M.; Gomes, R.; Mendonça, I.; Ferreira, A.; Cordeiro, N. Exopolymers from Cyanocohniella rudolphia: From modelling production to microplastics remediation applications. J. Clean. Prod. 2024, submitted.
- Jung, P.; Sommer, V.; Karsten, U.; Lakatos, M. Salt tolerance of terrestrial Cyanocohniella strains (Cyanobacteria) and description of C. rudolphia sp. nov. point to a marine origin of the genus and long-distance terrestrial dispersal patterns. Microorganisms 2022, 10, 968. [Google Scholar] [CrossRef]
- Cunha, C.; Faria, M.; Nogueira, N.; Ferreira, A.; Cordeiro, N. Marine vs freshwater microalgae exopolymers as biosolutions to microplastics pollution. Environ. Pollut. 2019, 249, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Babiak, W.; Krzemińska, I. Extracellular polymeric substances (EPS) as microalgal bioproducts: A Review of factors affecting eps synthesis and application in flocculation processes. Energies 2021, 14, 4007. [Google Scholar] [CrossRef]
- Gongi, W.; Pinchetti, J.L.G.; Cordeiro, N.; Sadok, S.; Ouada, H.B. Extracellular polymeric substances produced by the thermophilic cyanobacterium Gloeocapsa gelatinosa: Characterization and assessment of their antioxidant and metal-chelating activities. Mar. Drugs 2022, 20, 227. [Google Scholar] [CrossRef] [PubMed]
- Reignier, O.; Bormans, M.; Marchand, L.; Sinquin, C.; Amzil, Z.; Zykwinska, A.; Briand, E. Production and composition of extracellular polymeric substances by a unicellular strain and natural colonies of Microcystis: Impact of salinity and nutrient stress. Environ. Microbiol. Rep. 2023, 15, 783–796. [Google Scholar] [CrossRef] [PubMed]
- Gongi, W.; Cordeiro, N.; Pinchetti, J.L.G.; Sadok, S.; Ben Ouada, H. Extracellular polymeric substances with high radical scavenging ability produced in outdoor cultivation of the thermotolerant chlorophyte Graesiella sp. J. Appl. Phycol. 2021, 33, 357–369. [Google Scholar] [CrossRef]
- Laroche, C. Exopolysaccharides from microalgae and cyanobacteria: Diversity of strains, production strategies, and applications. Mar. Drugs 2022, 20, 336. [Google Scholar] [CrossRef] [PubMed]
- Morais, M.G.; Santos, T.D.; Moraes, L.; Vaz, B.S.; Morais, E.G.; Costa, J.A.V. Exopolysaccharides from microalgae: Production in a biorefinery framework and potential applications. Bioresour. Technol. Rep. 2022, 18, 101006. [Google Scholar] [CrossRef]
- Koçer, A.T.; İnan, B.; Kaptan Usul, S.; Koçer, B.; Çakmak, Y.S.; Usul, M.; Mendi, A.; Gencer, S. Exopolysaccharides from microalgae: Production, characterization, optimization and techno-economic assessment. Braz. J. Microbiol. 2021, 52, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Cunha, C.; Silva, L.; Paulo, J.; Faria, M.; Nogueira, N.; Cordeiro, N. Microalgal-based biopolymer for nano- and microplastic removal: A possible biosolution for wastewater treatment. Environ. Pollut. 2020, 263 Pt B, 114385. [Google Scholar] [CrossRef] [PubMed]

| Independent Variables | Units | Type of Variable | Variable Level * | ||
|---|---|---|---|---|---|
| −1 | 0 | +1 | |||
| Ratio (culture medium/wet biomass) (R) | mL/g | Continuous | 1:1 | 7.5:1 | 14:1 |
| Days (D) | days | Discrete | 1 | 4 | 7 |
| Nitrogen concentration (N) | g/L | Continuous | 2.50 | 6.25 | 10.00 |
| Phosphorus concentration (P) # | g/L | Continuous | 0.50 | 1.25 | 2.00 |
| N | Independent Variables—Production Conditions (Coded Levels) ^ | Response—S-EPS Production (mg/L) # | |||||
|---|---|---|---|---|---|---|---|
| Ratio (Culture Medium/Wet Biomass) (mL/g) | Production Days (D; day) | Phosphorus Concentration (P; g/L) | Nitrogen Concentration (N; g/L) | Experimental Values | Predicted Values | Variation (%) * | |
| 1 | 1:1 (−1) | 1 (−1) | 0.50 (−1) | 2.50 (−1) | 192.86 ± 8.55 | 199.52 | −3.45 |
| 2 | 1:1 (−1) | 1 (−1) | 0.50 (−1) | 10.00 (+1) | 217.92 ± 12.28 | 208.79 | 4.19 |
| 3 | 1:1 (−1) | 1 (−1) | 2.00 (+1) | 2.50 (−1) | 224.53 ± 5.66 | 225.52 | −0.44 |
| 4 | 1:1 (−1) | 1 (−1) | 2.00 (+1) | 10.00 (+1) | 228.65 ± 16.31 | 228.97 | −0.14 |
| 5 | 1:1 (−1) | 7 (+1) | 0.50 (−1) | 2.50 (−1) | 199.64 ± 63.96 | 186.90 | 6.38 |
| 6 | 1:1 (−1) | 7 (+1) | 0.50 (−1) | 10.00 (+1) | 178.69 ± 21.11 | 194.32 | −8.75 |
| 7 | 1:1 (−1) | 7 (+1) | 2.00 (+1) | 2.50 (−1) | 200.96 ± 14.04 | 203.96 | −1.49 |
| 8 | 1:1 (−1) | 7 (+1) | 2.00 (+1) | 10.00 (+1) | 206.82 ± 2.94 | 203.73 | 1.49 |
| 9 | 14:1 (+1) | 1 (−1) | 0.50 (−1) | 2.50 (−1) | 70.46 ± 8.82 | 67.56 | 4.12 |
| 10 | 14:1 (+1) | 1 (−1) | 0.50 (−1) | 10.00 (+1) | 112.49 ± 0.77 | 113.89 | −1.24 |
| 11 | 14:1 (+1) | 1 (−1) | 2.00 (+1) | 2.50 (−1) | 72.73 ± 8.52 | 61.51 | 15.43 |
| 12 | 14:1 (+1) | 1 (−1) | 2.00 (+1) | 10.00 (+1) | 93.44 ± 3.37 | 100.19 | −7.22 |
| 13 | 14:1 (+1) | 7 (+1) | 0.50 (−1) | 2.50 (−1) | 143.40 ± 8.69 | 149.33 | −4.14 |
| 14 | 14:1 (+1) | 7 (+1) | 0.50 (−1) | 10.00 (+1) | 200.78 ± 11.09 | 193.80 | 3.48 |
| 15 | 14:1 (+1) | 7 (+1) | 2.00 (+1) | 2.50 (−1) | 131.20 ± 5.19 | 134.34 | −2.39 |
| 16 | 14:1 (+1) | 7 (+1) | 2.00 (+1) | 10.00 (+1) | 173.40 ± 14.18 | 171.16 | 1.29 |
| 17 | 7.5:1 (0) | 4 (0) | 1.25 (0) | 6.25 (0) | 160.31 ± 15.66 | 142.34 | 11.21 |
| 18 | 1:1 (−1) | 4 (0) | 1.25 (0) | 6.25 (0) | 226.75 ± 1.55 | 205.61 | 9.32 |
| 19 | 7.5:1 (0) | 1 (−1) | 1.25 (0) | 6.25 (0) | 115.44 ± 10.65 | 128.88 | −11.64 |
| 20 | 7.5:1 (0) | 4 (0) | 0.50 (−1) | 6.25 (0) | 132.21 ± 1.17 | 143.34 | −8.42 |
| 21 | 7.5:1 (0) | 4 (0) | 1.25 (0) | 2.50 (−1) | 120.24 ± 8.22 | 129.03 | −7.31 |
| 22 | 7.5:1 (0) | 4 (0) | 2.00 (+1) | 6.25 (0) | 130.89 ± 4.07 | 142.34 | −8.75 |
| 23 | 7.5:1 (0) | 4 (0) | 1.25 (0) | 10.00 (+1) | 157.07 ± 2.98 | 155.65 | 0.90 |
| 25 | 7.5:1 (0) | 7 (+1) | 1.25 (0) | 6.25 (0) | 162.24 ± 5.61 | 155.80 | 3.97 |
| 25 | 14:1 (+1) | 4 (0) | 1.25 (0) | 6.25 (0) | 135.24 ± 6.75 | 125.91 | 6.90 |
| Sum of Squares | Degree of Freedom | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|
| Model | 1.59 × 105 | 14 | 11,361.83 | 40.06 | <0.0001 |
| R | 92,171.23 | 1 | 92,171.23 | 325.02 | <0.0001 |
| D | 12,026.63 | 1 | 12,026.63 | 42.41 | <0.0001 |
| P | 33.53 | 1 | 33.53 | 0.12 | 0.7321 |
| N | 7578.77 | 1 | 7578.77 | 26.72 | <0.0001 |
| R*D | 26,725.88 | 1 | 26,725.88 | 94.24 | <0.0001 |
| R*P | 3082.14 | 1 | 3082.14 | 10.87 | 0.0016 |
| R*N | 4119.63 | 1 | 4119.63 | 14.53 | 0.0003 |
| D*P | 239.76 | 1 | 239.76 | 0.85 | 0.3614 |
| D*N | 10.33 | 1 | 10.33 | 0.04 | 0.8492 |
| P*N | 175.79 | 1 | 175.79 | 0.62 | 0.4340 |
| R*R | 11,017.20 | 1 | 11,017.20 | 38.85 | <0.0001 |
| D*D | 141.23 | 1 | 141.23 | 0.50 | 0.4830 |
| P*P | 1029.33 | 1 | 1029.33 | 3.63 | 0.0613 |
| N*N | 153.57 | 1 | 153.57 | 0.54 | 0.4645 |
| Residual | 17,865.95 | 63 | 283.59 | ----- | ----- |
| Lack of Fit | 4834.92 | 10 | 483.49 | 1.97 | 0.0561 |
| Error | 13,031.03 | 53 | 245.87 | ----- | ----- |
| N | Independent Variables—Production Conditions (Coded Levels) ^ | Response—S-EPS Production (mg/L) # | ||||
|---|---|---|---|---|---|---|
| Ratio (Culture Medium/Wet Biomass) (mL/g) | Production Days (D; day) | Nitrogen Concentration (N; g/L) | Experimental Values | Predicted Values | Variation (%) * | |
| 1 | 1:1 (−1) | 1 (−1) | 2.50 (−1) | 211.70 ± 9.41 | 212.47 | −0.36 |
| 2 | 1:1 (−1) | 1 (−1) | 10.00 (+1) | 219.65 ± 1.19 | 218.56 | 0.50 |
| 3 | 1:1 (−1) | 4 (0) | 6.25 (0) | 200.49 ± 8.49 | 222.09 | −10.77 |
| 4 | 1:1 (−1) | 7 (+1) | 2.50 (−1) | 197.60 ± 8.49 | 196.05 | 0.78 |
| 5 | 1:1 (−1) | 7 (+1) | 10.00 (+1) | 205.42 ± 7.06 | 200.29 | 2.50 |
| 6 | 7.5:1 (0) | 1 (−1) | 6.25 (0) | 142.06 ± 1.79 | 123.70 | 12.92 |
| 7 | 7.5:1 (0) | 4 (0) | 2.50 (−1) | 128.03 ± 1.20 | 126.58 | 1.13 |
| 8 | 7.5:1 (0) | 4 (0) | 6.25 (0) | 149.15 ± 14.00 | 146.15 | 2.01 |
| 9 | 7.5:1 (0) | 4 (0) | 10.00 (+1) | 154.73 ± 3.56 | 150.28 | 2.88 |
| 10 | 7.5:1 (0) | 7 (+1) | 6.25 (0) | 155.86 ± 2.50 | 153.54 | 1.49 |
| 11 | 14:1 (+1) | 1 (−1) | 2.50 (−1) | 65.57 ± 4.44 | 64.12 | 2.21 |
| 12 | 14:1 (+1) | 7 (+1) | 2.50 (−1) | 138.18 ± 1.54 | 142.08 | −2.82 |
| 13 | 14:1 (+1) | 1 (−1) | 10.00 (+1) | 104.80 ± 8.88 | 107.27 | −2.36 |
| 14 | 14:1 (+1) | 7 (+1) | 10.00 (+1) | 183.01 ± 5.10 | 183.38 | −0.20 |
| 15 | 14:1 (+1) | 4 (0) | 6.25 (0) | 136.31 ± 4.76 | 139.46 | −2.31 |
| Sum of Squares | Degree of Freedom | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|
| Model | 1.467 × 105 | 6 | 24,450.32 | 92.48 | <0.0001 |
| R | 83,999.52 | 1 | 83,999.52 | 317.72 | <0.0001 |
| D | 9583.41 | 1 | 9583.41 | 36.25 | <0.0001 |
| N | 9370.07 | 1 | 9370.07 | 35.44 | <0.0001 |
| R*D | 29,880.44 | 1 | 29,880.44 | 113.02 | <0.0001 |
| R*N | 2721.69 | 1 | 2721.69 | 10.29 | 0.0020 |
| R*R | 9050.72 | 1 | 9050.72 | 34.23 | <0.0001 |
| Residual | 18,506.94 | 70 | 264.38 | ||
| Lack of Fit | 3586.34 | 8 | 448.29 | 1.86 | 0.0823 |
| Error | 14,920.61 | 62 | 240.65 |
| General Model (Equation (4)) | Simplified Model (Equation (5)) | |
|---|---|---|
| Predicted | 0.9092 | 0.8629 |
| Adjusted | 0.9285 | 0.8784 |
| 0.9417 | 0.8880 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, F.; Mendonça, I.; Faria, M.; Gomes, R.; Pinchetti, J.L.G.; Ferreira, A.; Cordeiro, N. Response Surface Methodology Applied to Cyanobacterial EPS Production: Steps and Statistical Validations. Processes 2024, 12, 1733. https://doi.org/10.3390/pr12081733
Rodrigues F, Mendonça I, Faria M, Gomes R, Pinchetti JLG, Ferreira A, Cordeiro N. Response Surface Methodology Applied to Cyanobacterial EPS Production: Steps and Statistical Validations. Processes. 2024; 12(8):1733. https://doi.org/10.3390/pr12081733
Chicago/Turabian StyleRodrigues, Filipa, Ivana Mendonça, Marisa Faria, Ricardo Gomes, Juan Luis Gómez Pinchetti, Artur Ferreira, and Nereida Cordeiro. 2024. "Response Surface Methodology Applied to Cyanobacterial EPS Production: Steps and Statistical Validations" Processes 12, no. 8: 1733. https://doi.org/10.3390/pr12081733
APA StyleRodrigues, F., Mendonça, I., Faria, M., Gomes, R., Pinchetti, J. L. G., Ferreira, A., & Cordeiro, N. (2024). Response Surface Methodology Applied to Cyanobacterial EPS Production: Steps and Statistical Validations. Processes, 12(8), 1733. https://doi.org/10.3390/pr12081733










