The Application of Fourier Transform Infrared Spectroscopy and Chemometrics in Identifying Signatures for Sheep’s Milk Authentication
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Material
2.2. Apparatus
2.3. Statistical and Chemometric Methods
2.4. Machine Learning for Milk Authentication
3. Results and Discussion
- (1)
- Exploratory analysis;
- (2)
- Principal Component Analysis (PCA);
- (3)
- Regression task—to predict intensity based on wavelengths;
- (4)
- Classification task—to find the milk type based on parameters analysis.
3.1. Exploratory Analysis
3.2. Principal Component Analysis (PCA)
3.3. Regression Task
- Detection of cow’s milk in bryndza using the HPLC-DAD-MS method—a relatively expensive approach due to the need for costly, technically advanced equipment and the relatively complex process of sample preparation for analysis and extraction [3].
- Identification of volatile compounds and fatty acids characteristic of different types of bryndza using GC/MS [13].
- Non-traditional but highly economical methods for identifying the origin of bryndza and milk or detecting the adulteration of 100% sheep’s milk bryndza with cow’s milk cheeses, such as FTIR analysis, which was the focus of this project. The equipment itself may not be the cheapest, but it allows for quick and easy processing with minimal interference with the matrix, which is a clear advantage of this approach [16].
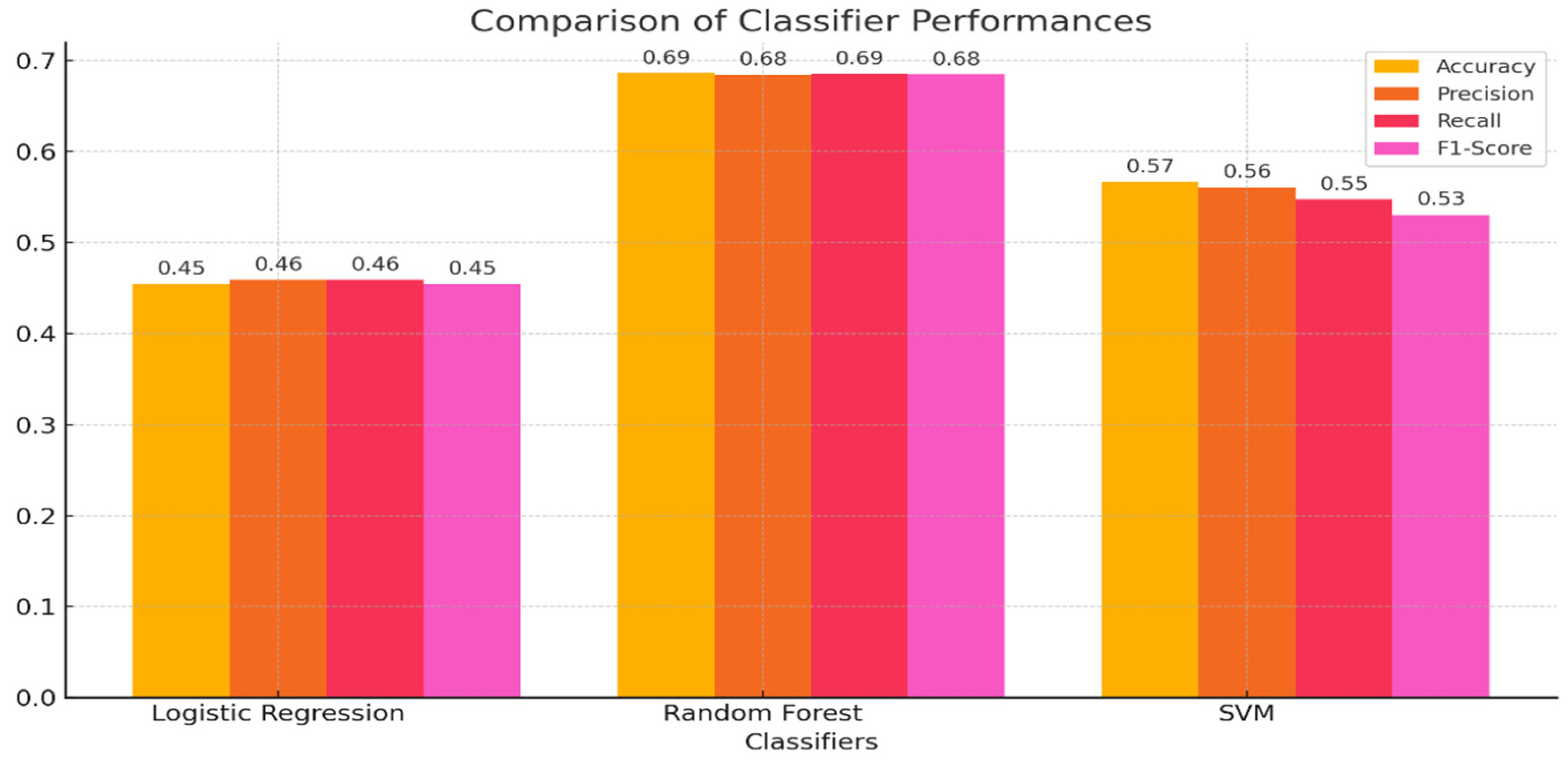
3.4. Classification Task
- Logistic Regression: Regularization parameter (C);
- Random Forest: Number of trees (n_estimators) and maximum depth (max_depth);
- SVM: Kernel type and regularization.
- ○
- Best Parameter: C = 10
- ○
- Best Cross-Validation Score: 52.4%
- ○
- Best Parameters: max_depth = 20, n_estimators = 50
- ○
- Best Cross-Validation Score: 75.3%
- ○
- Best Parameters: C = 10, kernel = ‘rbf’
- ○
- Best Cross-Validation Score: 57.7%
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grunert, K.G.; Aachmann, K. Consumer reactions to the use of EU quality labels on food products: A review of the literature. Food Control 2016, 59, 178–187. [Google Scholar] [CrossRef]
- Dias, C.; Mendes, L. Protected Designation of Origin (PDO), Protected Geographical Indication (PGI) and Traditional Speciality Guaranteed (TSG): A bibliometric analysis. Food Res. Int. 2018, 103, 492–508. [Google Scholar] [CrossRef] [PubMed]
- González-Domínguez, R. Food Authentication: Techniques, Trends and Emerging Approaches (Second Issue). Foods 2022, 11, 1926. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Wu, B.; Zhu, S.; He, Y.; Zhang, C. Application of Visible/Infrared Spectroscopy and Hyperspectral Imaging with Machine Learning Techniques for Identifying Food Varieties and Geographical Origins. Front. Nutr. 2021, 8, 680357. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Song, H.; Guo, L.; Guang, P.; Yang, X.; Li, L.; Zhao, H.; Yang, M. Detection of adulteration in Chinese honey using NIR and ATR-FTIR spectral data fusion. Spectrochim. Acta. Part A Mol. Biomol. Spectrosc. 2020, 235, 118297. [Google Scholar] [CrossRef] [PubMed]
- Taylan, O.; Cebi, N.; Yilmaz, M.T.; Sagdic, O.; Ozdemir, D.; Balubaid, M. Rapid detection of green-pea adulteration in pistachio nuts using Raman spectroscopy and chemometrics. J. Sci. Food Agric. 2021, 101, 1699–1708. [Google Scholar] [CrossRef] [PubMed]
- Didham, M.; Truong, V.K.; Chapman, J.; Cozzolino, D. Sensing the Addition of Vegetable Oils to Olive Oil: The Ability of UV–VIS and MIR Spectroscopy Coupled with Chemometric Analysis. Food Anal. Methods 2020, 13, 601–607. [Google Scholar] [CrossRef]
- Cubero-Leon, E.; Peñalver, R.; Maquet, A. Review on metabolomics for food authentication. Food Res. Int. 2014, 60, 95–107. [Google Scholar] [CrossRef]
- Nallan Chakravartula, S.S.; Moscetti, R.; Bedini, G.; Nardella, M.; Massantini, R. Use of convolutional neural network (CNN) combined with FT-NIR spectroscopy to predict food adulteration: A case study on coffee. Food Control 2022, 135, 108816. [Google Scholar] [CrossRef]
- Leo, M.; Sharma, S.; Maddulety, K. Machine learning in banking risk management: A literature review. Risks 2019, 7, 29. [Google Scholar] [CrossRef]
- Fuentes, S.; Torrico, D.D.; Tongson, E.; Viejo, C.G. Machine learning modeling of wine sensory profiles and color of vertical vintages of pinot noir based on chemical fingerprinting, weather and management data. Sensors 2020, 20, 3618. [Google Scholar] [CrossRef] [PubMed]
- Haque, E.; Taniguchi, H.; Hassan, M.M.; Bhowmik, P.; Karim, M.R.; Śmiech, M.; Zhao, K.; Rahman, M.; Islam, T. Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: Recent progress, prospects, and challenges. Front. Plant Sci. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Xiong, J.; Chen, S.; Yu, H.; Chen, C.; Huang, J.; Yuan, H.; Lou, H. Rapid identification of adulteration in raw bovine milk with soymilk by electronic nose and headspace-gas chromatography ion-mobility spectrometry. Food Chem. 2023, 18, 100696. [Google Scholar] [CrossRef] [PubMed]
- ISO 13366-2:2006; Milk—Enumeration of Somatic Cells. International Organization for Standardization (ISO): Geneva, Switzerland, 2006.
- ISO 9622:2013; Milk and Liquid Milk Products—Guidelines for the Application of Mid-Infrared Spectrometry. International Organization for Standardization (ISO): Geneva, Switzerland, 2013.
- Sen, S.; Dundar, Z.; Uncu, O.; Ozen, B. Potential of Fourier-transform infrared spectroscopy in adulteration detection and quality assessment in buffalo and goat milks. Microchem. J. 2021, 166, 106207. [Google Scholar] [CrossRef]
- Gantner, V.; Mijić, P.; Baban, M.; Škrtić, Z.; Turalija, A. The overall and fat composition of milk of various species. Mljekarstvo 2015, 65, 223–231. [Google Scholar] [CrossRef]
- Roy, D.; Ye, A.; Moughan, P.J. Singh Harjinder: Composition, Structure, and Digestive Dynamics of Milk from Different Species—A Review. Front. Nutr. 2020, 7, 577759. [Google Scholar] [CrossRef] [PubMed]
- Cirak, O.; Icyer, N.C.; Durak, M.Z. Rapid detection of adulteration of milks from different species using Fourier Transform Infrared Spectroscopy (FTIR). J. Dairy Res. 2018, 85, 222–225. [Google Scholar] [CrossRef] [PubMed]
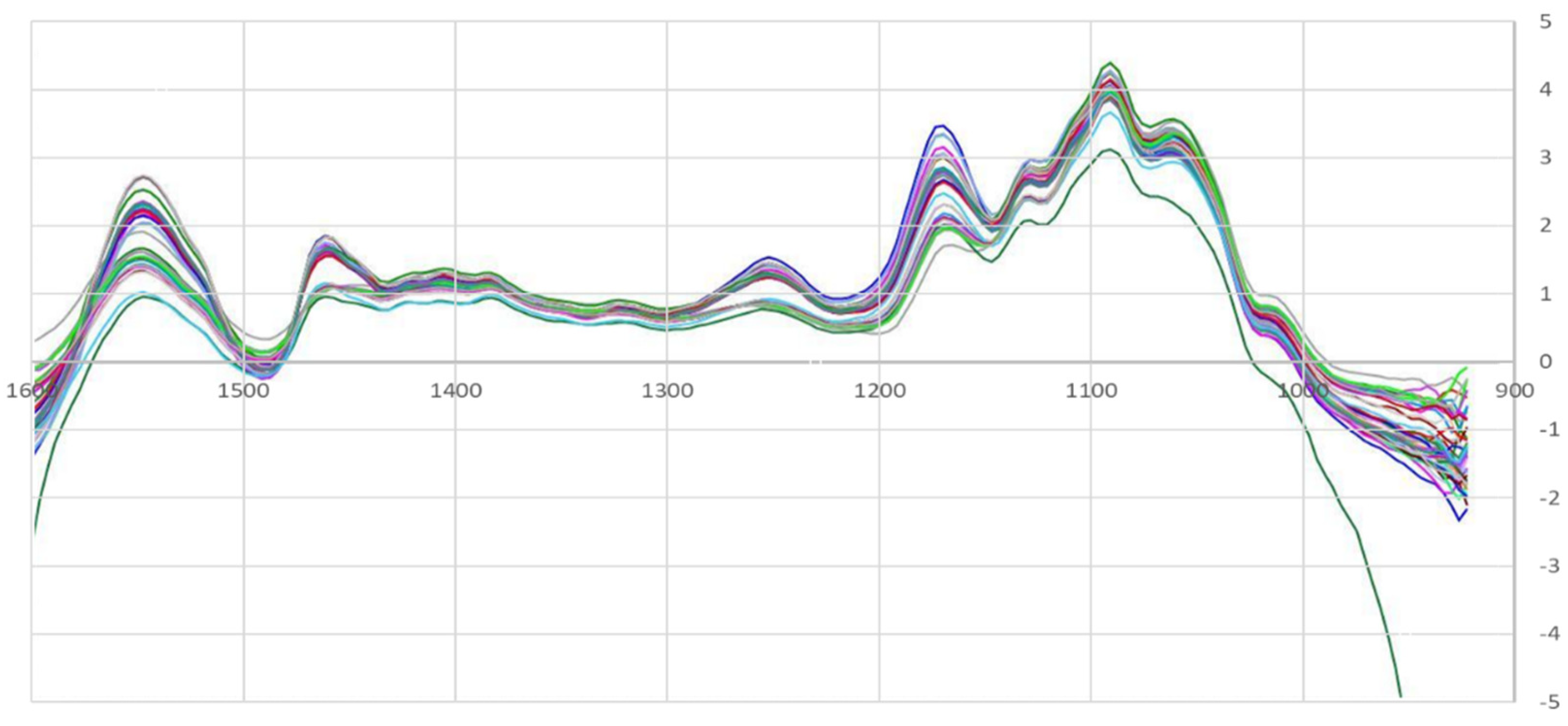
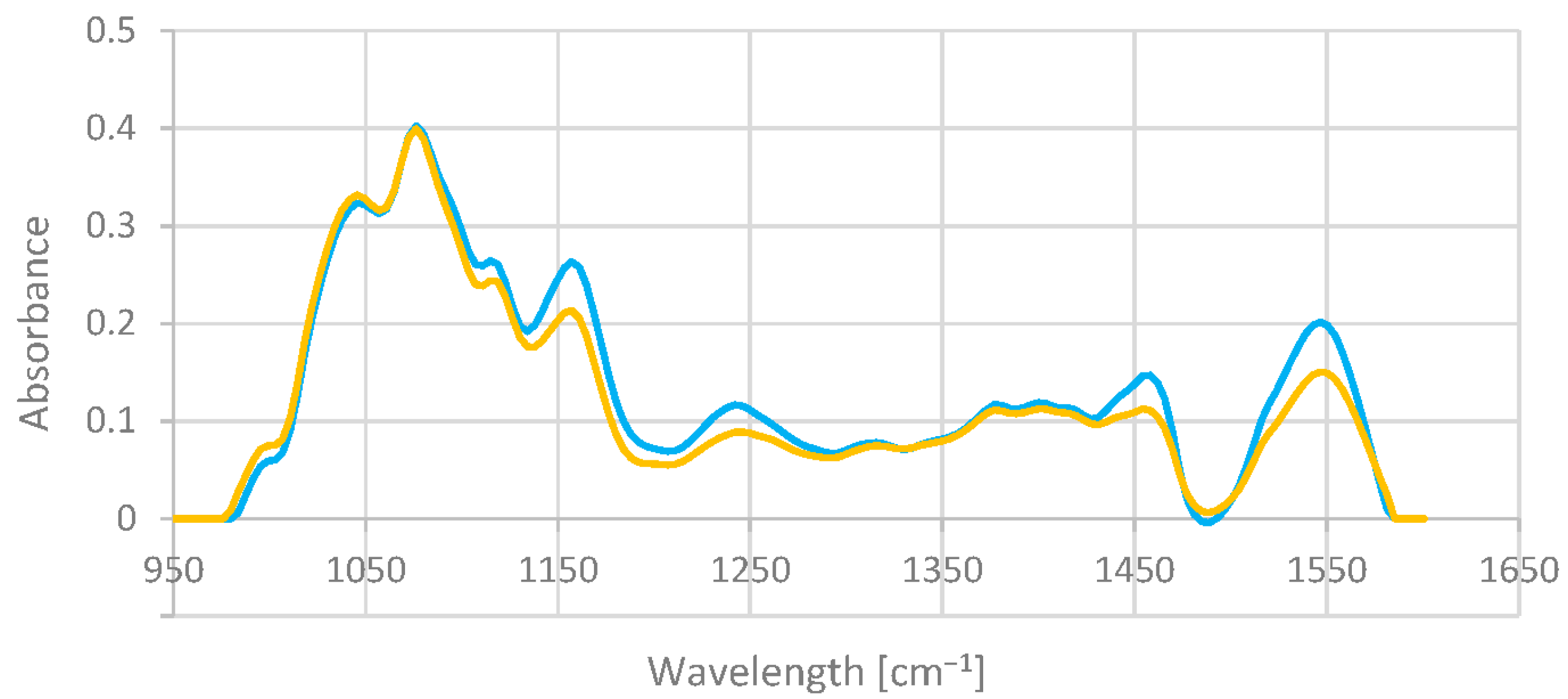
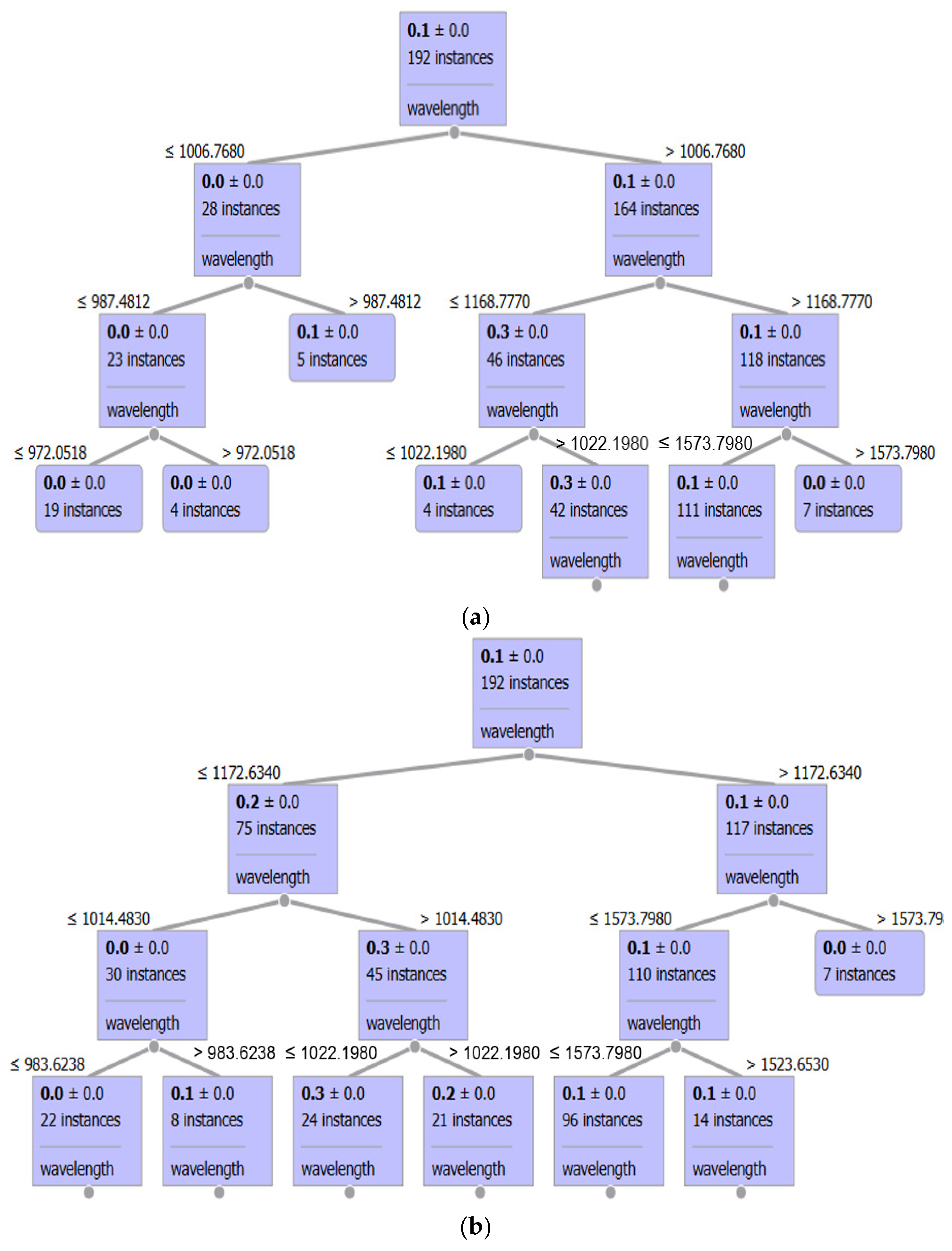
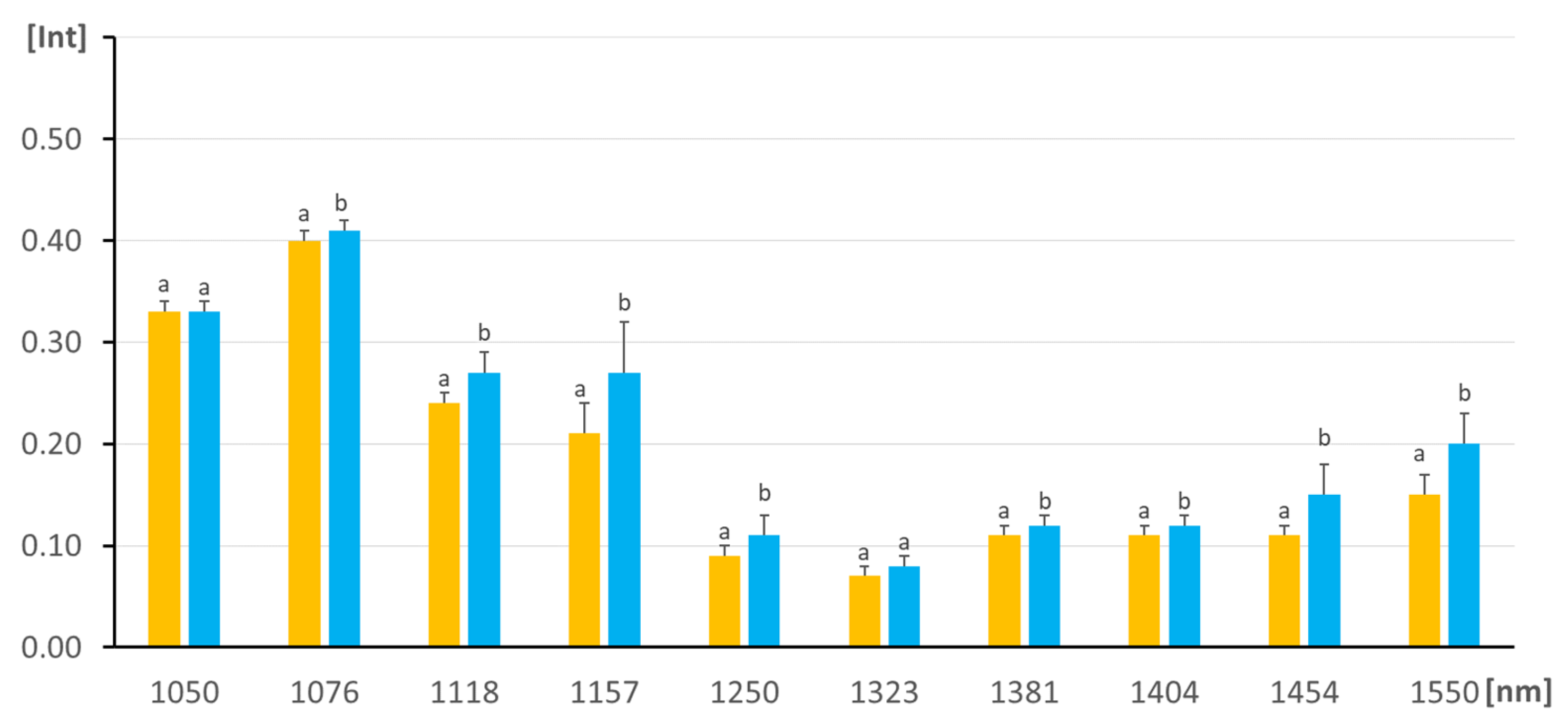
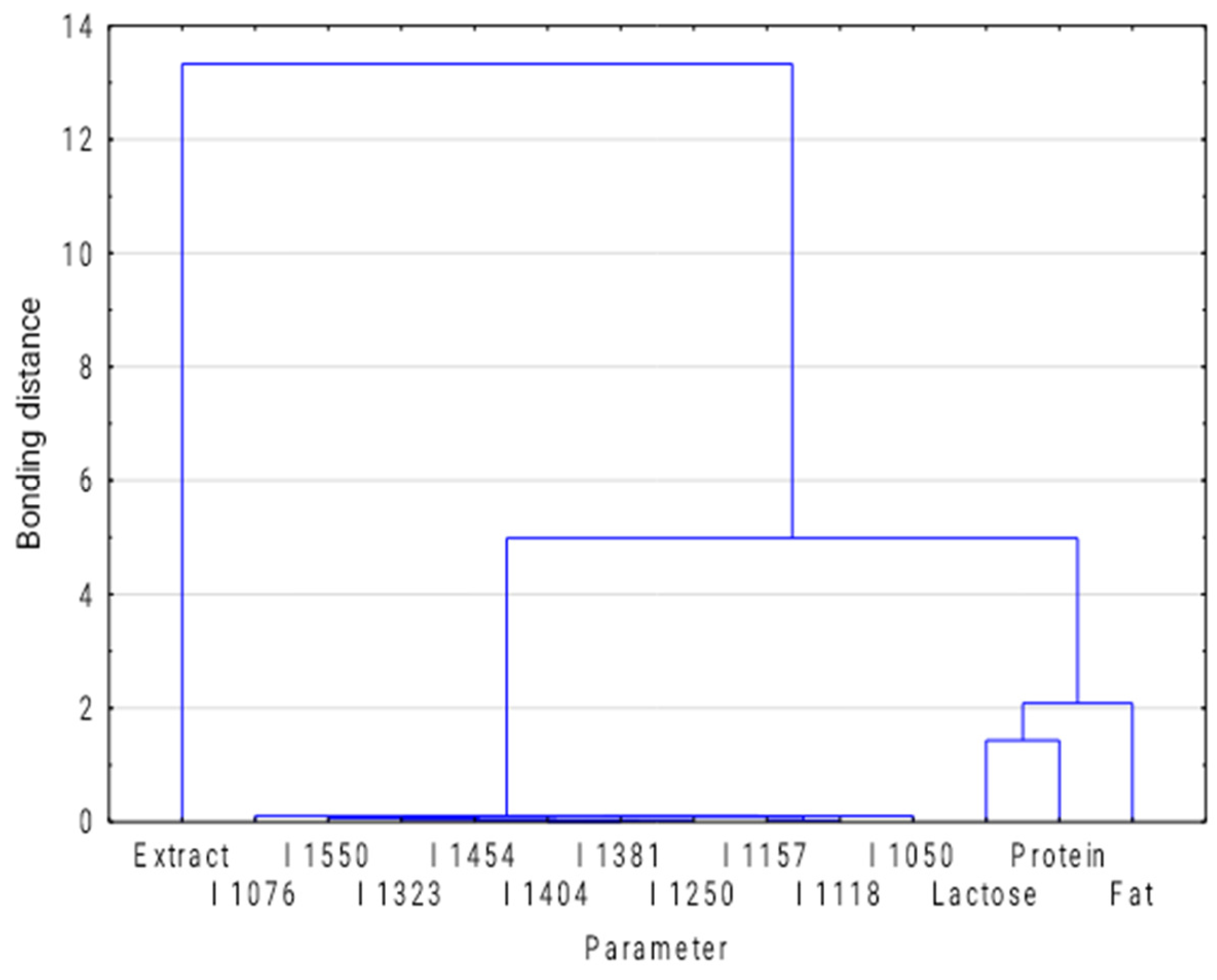
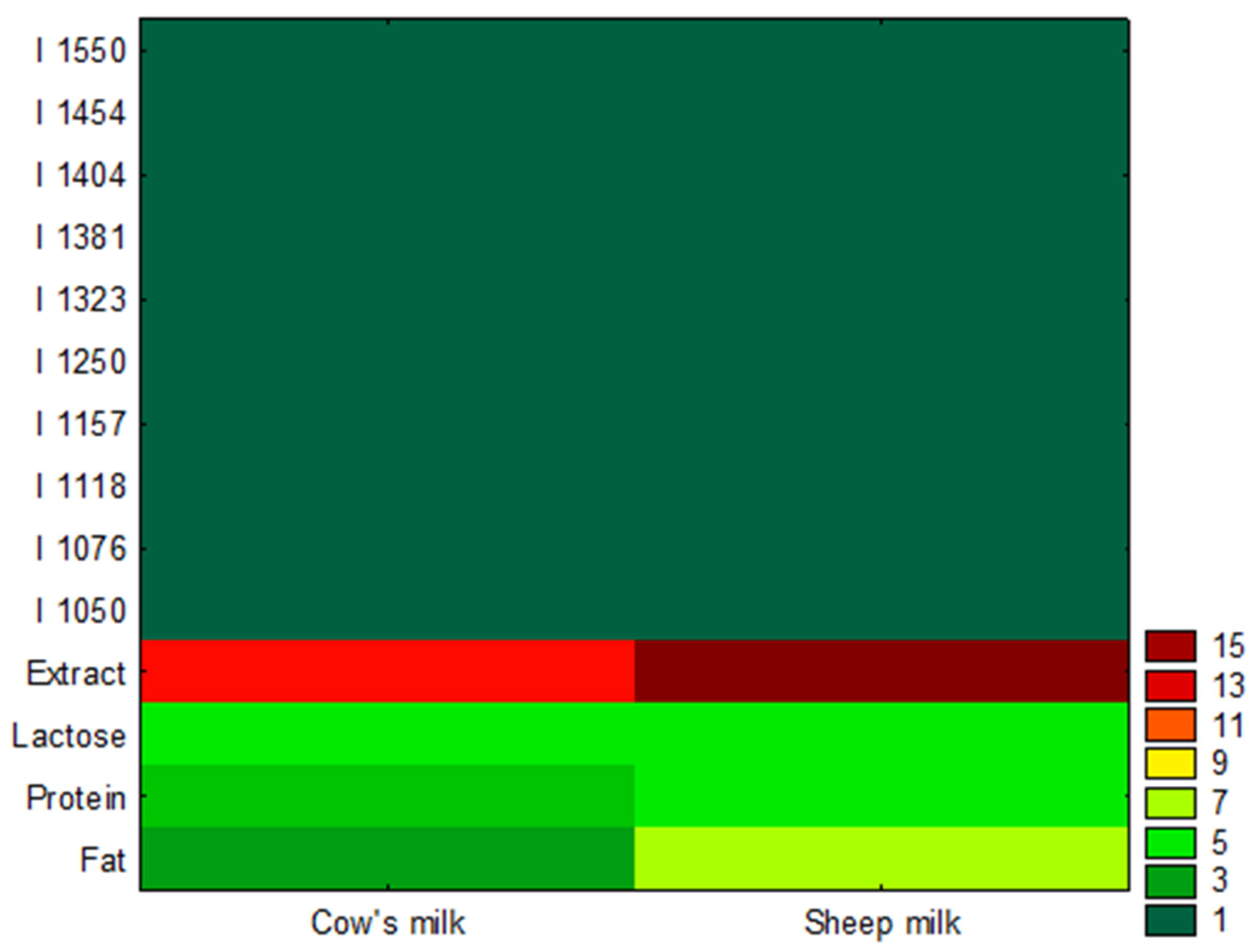
| Sample | Fat (% m/m) | SD | Protein (% m/m) | SD | Lactose (% m/m) | SD | Extract (% m/m) | SD |
|---|---|---|---|---|---|---|---|---|
| Cow’s milk | 2.81 a | 1.28 | 3.30 a | 0.08 | 4.73 a | 0.14 | 11.52 a | 1.26 |
| Sheep’s milk | 6.49 b | 2.98 | 4.47 b | 1.23 | 4.56 b | 0.39 | 16.59 b | 3.36 |
| Parameter | Fat | Protein | Lactose | Extract | I 1050 | I 1076 | I 1118 | I 1157 | I 1250 | I 1323 |
|---|---|---|---|---|---|---|---|---|---|---|
| PC1 | −1.0 | −1.0 | 1.0 | −1.0 | −1.0 | −1.0 | −1.0 | −1.0 | −1.0 | −1.0 |
| Milk Type | PC1 |
|---|---|
| Cow | 2.64575 |
| Sheep | −2.64575 |
| Dataset Type | Model | R2 |
|---|---|---|
| Sheep | Decision tree | 0.9766 |
| Random forest | 0.9778 | |
| Linear regression | 0.0188 | |
| Support vector machine with a linear kernel | −0.828 | |
| Support vector machine with a polynomial kernel | 0.027 | |
| Support vector machine with a radial-basis function | 0.621 | |
| KNN with k = 3 | 0.965 | |
| KNN with k = 5 | 0.961 | |
| KNN with k = 7 | 0.944 | |
| Cow | Decision tree | 0.9798 |
| Random forest | 0.9801 | |
| Linear regression | 0.0415 | |
| Support vector machine with a linear kernel | −0.102 | |
| Support vector machine with a polynomial kernel | 0.056 | |
| Support vector machine with a radial-basis function | 0.603 | |
| KNN with k = 3 | 0.970 | |
| KNN with k = 5 | 0.966 | |
| KNN with k = 7 | 0.951 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duliński, R.; Gancarz, M.; Shakhovska, N.; Byczyński, Ł. The Application of Fourier Transform Infrared Spectroscopy and Chemometrics in Identifying Signatures for Sheep’s Milk Authentication. Processes 2025, 13, 518. https://doi.org/10.3390/pr13020518
Duliński R, Gancarz M, Shakhovska N, Byczyński Ł. The Application of Fourier Transform Infrared Spectroscopy and Chemometrics in Identifying Signatures for Sheep’s Milk Authentication. Processes. 2025; 13(2):518. https://doi.org/10.3390/pr13020518
Chicago/Turabian StyleDuliński, Robert, Marek Gancarz, Nataliya Shakhovska, and Łukasz Byczyński. 2025. "The Application of Fourier Transform Infrared Spectroscopy and Chemometrics in Identifying Signatures for Sheep’s Milk Authentication" Processes 13, no. 2: 518. https://doi.org/10.3390/pr13020518
APA StyleDuliński, R., Gancarz, M., Shakhovska, N., & Byczyński, Ł. (2025). The Application of Fourier Transform Infrared Spectroscopy and Chemometrics in Identifying Signatures for Sheep’s Milk Authentication. Processes, 13(2), 518. https://doi.org/10.3390/pr13020518








