Antimicrobial and Anti-Biofilm Activities of Medicinal Plant-Derived Honey Against ESKAPE Pathogens: Insights into β-Lactamase Inhibition via Metabolomics and Molecular Modeling Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Honey Preparation and Collection
2.2. 1H-NMR Analysis
2.3. LC-MS Metabolomic Analysis
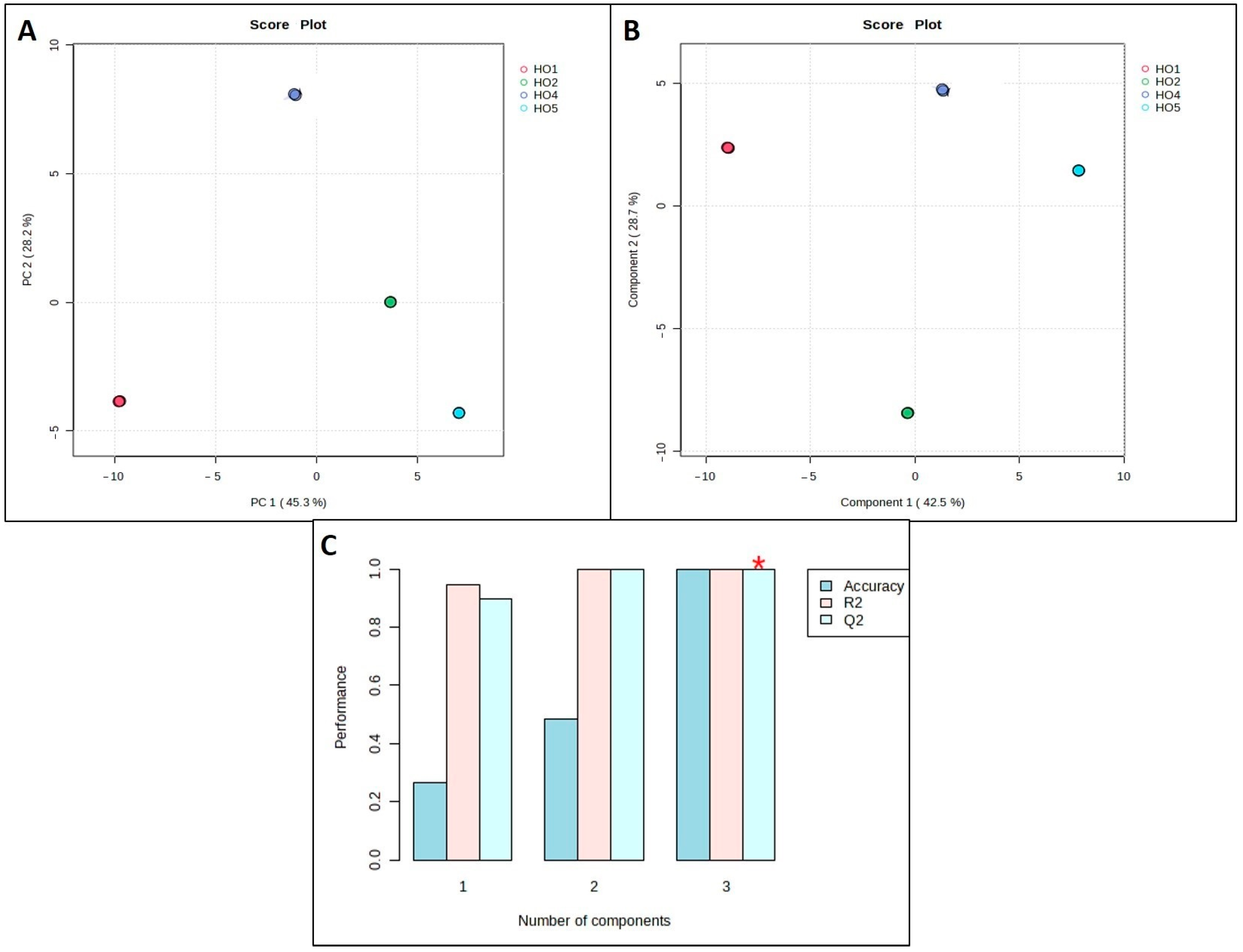
2.4. Multivariate Data Statistical Analysis
2.5. Antibacterial Activity
2.6. Biofilm Inhibition Assay
2.7. Molecular Docking
3. Results
3.1. Physicochemical Properties
3.2. Chemical Profiling
3.2.1. NMR Analysis
3.2.2. Multivariate Data Statistical Analysis
3.3. Antibacterial Activity of Saudi Multi-Floral Honey Samples
3.4. Anti-Biofilm Activity
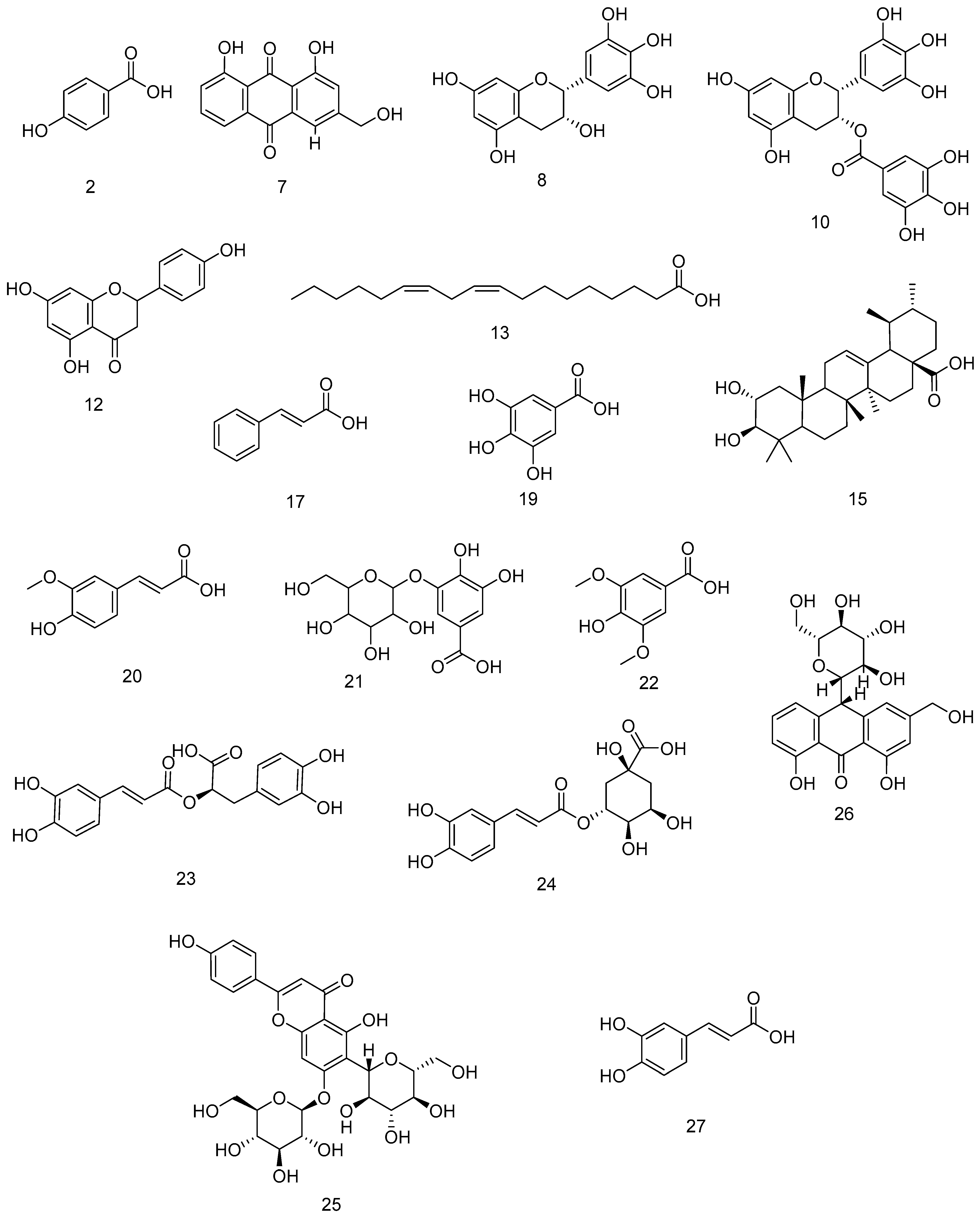
3.5. LC-MS Metabolomic Profiling
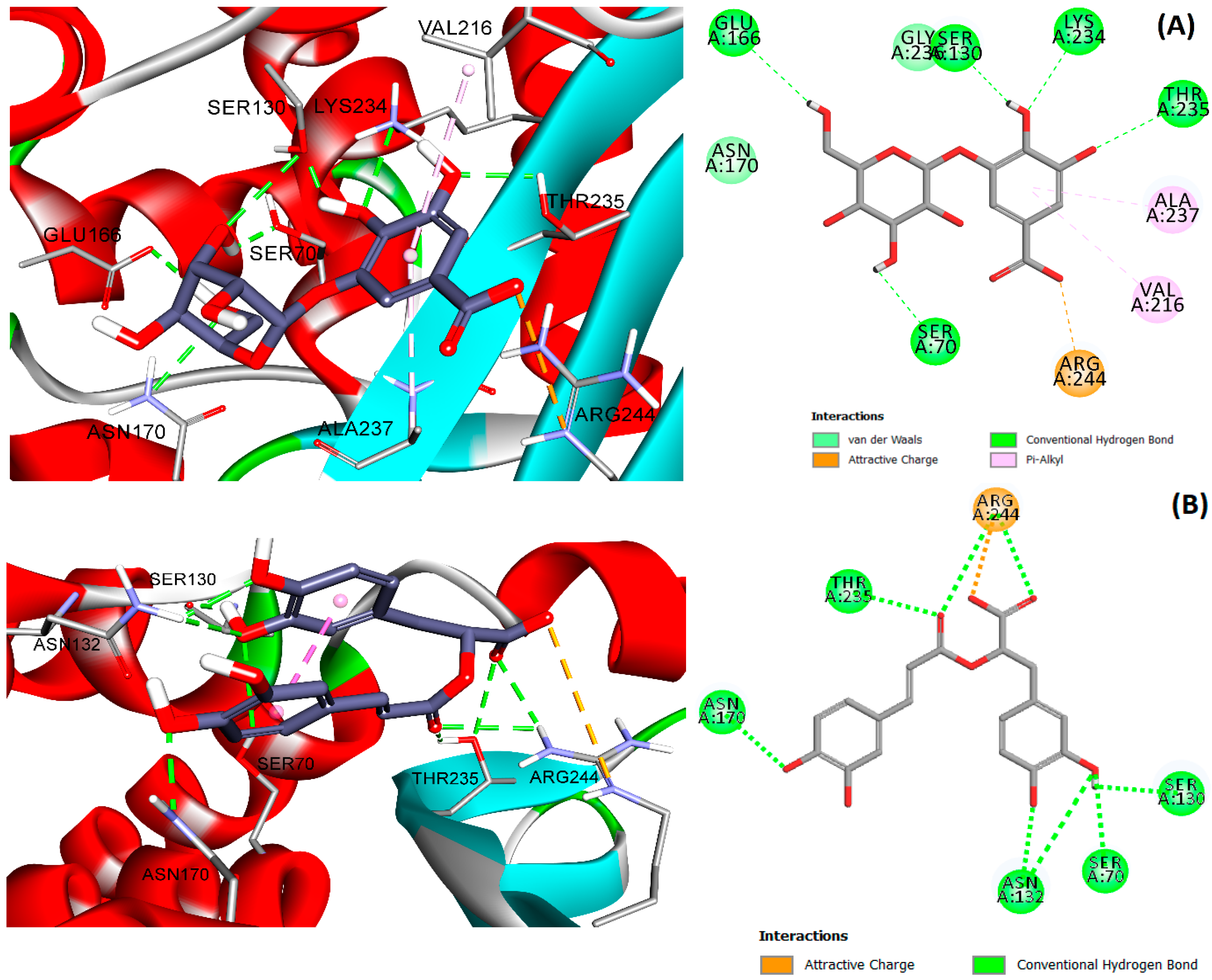
3.6. Molecular Docking
4. Discussion
4.1. Multivariate Data Analysis
4.2. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jadimurthy, R.; Mayegowda, S.B.; Nayak, S.C.; Mohan, C.D.; Rangappa, K.S. Escaping mechanisms of ESKAPE pathogens from antibiotics and their targeting by natural compounds. Biotechnol. Rep. 2022, 34, e00728. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Ngo, T.; Farooq, U.; Abagyan, R. Analysis of drug binding pockets and repurposing opportunities for twelve essential enzymes of ESKAPE pathogens. PeerJ 2017, 5, e3765. [Google Scholar] [CrossRef] [PubMed][Green Version]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Sarkar, J.; Singh, R.; Chandel, S. Understanding LC/MS-Based Metabolomics: A Detailed Reference for Natural Product Analysis. Proteom. Clin. Appl. 2025, 19, e202400048. [Google Scholar] [CrossRef]
- El-Hawary, S.S.; Mohammed, R.; Bahr, H.S.; Attia, E.Z.; El-Katatny, M.H.; Abelyan, N.; Al-Sanea, M.M.; Moawad, A.S.; Abdelmohsen, U.R. Soybean-associated endophytic fungi as potential source for anti-COVID-19 metabolites supported by docking analysis. J. Appl. Microbiol. 2021, 131, 1193–1211. [Google Scholar] [CrossRef]
- Kumar, D.; Hazra, K.; Prasad, P.V.V.; Bulleddu, R. Honey: An important nutrient and adjuvant for maintenance of health and management of diseases. J. Ethn. Foods 2024, 11, 19. [Google Scholar] [CrossRef]
- Hajian-Tilaki, A.; Kenari, R.E.; Razavi, R.; Farahmandfar, R. Phenolic profile, antioxidant properties, and pollen spectra of Iranian-originated honeys. Eur. Food Res. Technol. 2024, 250, 2317–2329. [Google Scholar] [CrossRef]
- Abdelhady, A.S.M.; Darwish, N.M.; Abdel-Rahman, S.M.; El Magd, N.M.A. The combined antimicrobial activity of citrus honey and fosfomycin on multidrug resistant pseudomonas aeruginosa isolates. AIMS Microbiol. 2020, 6, 162–175. [Google Scholar] [CrossRef]
- Tsadila, C.; Nikolaidis, M.; Dimitriou, T.G.; Kafantaris, I.; Amoutzias, G.D.; Pournaras, S.; Mossialos, D. Antibacterial activity and characterization of bacteria isolated from diverse types of greek honey against nosocomial and foodborne pathogens. Appl. Sci. 2021, 11, 5801. [Google Scholar] [CrossRef]
- Raheem, D.J.; Tawfike, A.F.; Abdelmohsen, U.R.; Edrada-Ebel, R.A.; Fitzsimmons-Thoss, V. Application of metabolomics and molecular networking in investigating the chemical profile and antitrypanosomal activity of British bluebells (Hyacinthoides non-scripta). Sci. Rep. 2019, 9, 2547. [Google Scholar] [CrossRef]
- Demarque, D.P.; Dusi, R.G.; de Sousa, F.D.M.; Grossi, S.M.; Silvério, M.R.S.; Lopes, N.P.; Espindola, L.S. Mass spectrometry-based metabolomics approach in the isolation of bioactive natural products. Sci. Rep. 2020, 10, 1051. [Google Scholar] [CrossRef] [PubMed]
- Alhadrami, H.A.; Sayed, A.M.; Sharif, A.M.; Azhar, E.I.; Rateb, M.E. Olive-derived triterpenes suppress SARS-CoV-2 main protease: A promising scaffold for future therapeutics. Molecules 2021, 26, 2654. [Google Scholar] [CrossRef] [PubMed]
- Attea, S.A.; Ghareeb, M.A.; Kelany, A.K.; Elhakim, H.K.A.; Allemailem, K.S.; Bukhari, S.I.; Rashidi, F.B.; Hamed, A.A. Biosynthesis of Iron Oxide Nanoparticles by Marine Streptomyces sp. SMGL39 with Antibiofilm Activity: In Vitro and In Silico Study. Molecules 2024, 29, 4784. [Google Scholar] [CrossRef] [PubMed]
- Khanfar, M.A.; Taha, M.O. Elaborate ligand-based modeling coupled with multiple linear regression and k nearest neighbor QSAR analyses unveiled new nanomolar mTOR inhibitors. J. Chem. Inf. Model. 2013, 53, 2587–2612. [Google Scholar] [CrossRef]
- Khanfar, M.A.; Salaas, N.; Abumostafa, R. Discovery of natural-derived Mpro inhibitors as therapeutic candidates for COVID-19: Structure-based pharmacophore screening combined with QSAR analysis. Mol. Inform. 2023, 42, 2200198. [Google Scholar] [CrossRef]
- Nandigrami, P.; Fiser, A. Assessing the functional impact of protein binding site definition. Protein Sci. 2024, 33, e5026. [Google Scholar] [CrossRef]
- Ricochon, G.; Paris, C.; Girardin, M.; Muniglia, L. Highly sensitive, quick and simple quantification method for mono and disaccharides in aqueous media using liquid chromatography–atmospheric pressure chemical ionization–mass spectrometry (LC–APCI–MS). J. Chromatogr. B 2011, 879, 1529–1536. [Google Scholar] [CrossRef]
- Wang, Y.; Xing, L.; Zhang, J.; Ma, X.; Weng, R. Determination of endogenous phenolic compounds in honey by HPLC-MS/MS. LWT 2023, 183, 114951. [Google Scholar] [CrossRef]
- Li, T.; Zhao, T. Analysis of Raffinose Metabolism-Related Sugar Components in Seeds. In Seed Dormancy: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2024; pp. 73–80. [Google Scholar]
- Soyseven, M.; Sezgin, B.; Arli, G. A novel, rapid and robust HPLC-ELSD method for simultaneous determination of fructose, glucose and sucrose in various food samples: Method development and validation. J. Food Compos. Anal. 2022, 107, 104400. [Google Scholar] [CrossRef]
- Zhou, J.; Qi, Y.; Ritho, J.; Duan, L.; Wu, L.; Diao, Q.; Li, Y.; Zhao, J. Analysis of maltooligosaccharides in honey samples by ultra-performance liquid chromatography coupled with evaporative light scattering detection. Food Res. Int. 2014, 56, 260–265. [Google Scholar] [CrossRef]
- Valverde, S.; Ares, A.M.; Elmore, J.S.; Bernal, J. Recent trends in the analysis of honey constituents. Food Chem. 2022, 387, 132920. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Abdulla, R.; Ma, Q.; Aisa, H.A. Comprehensive Identification of Chemical Fingerprint and Screening of Potential Quality Markers of Aloe vera (L.) Burm. f. from Different Geographical Origins via Ultra-High-Performance Liquid Chromatography Hyphenated with Quadrupole–Orbitrap-High-Resolu. J. Chromatogr. Sci. 2023, 61, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Susanti, E.; Ratnawati, R.; Rudijanto, A. Qualitative analysis of catechins from green tea GMB-4 clone using HPLC and LC-MS/MS. Asian Pac. J. Trop. Biomed. 2015, 5, 1046–1050. [Google Scholar] [CrossRef]
- Sorucu, A.; Oruç, H.H. Determination of biologically active phenolic compounds in propolis by LC–MS/MS according to seasons and altitudes. J. Food Meas. Charact. 2019, 13, 2461–2469. [Google Scholar] [CrossRef]
- An, N.; Cai, W.-J.; Zhu, Q.-F.; Wang, W.; Hussain, D.; Feng, Y.-Q. Metabolic profiling of organic acids in honey by stable isotope labeling assisted liquid chromatography-mass spectrometry. J. Food Compos. Anal. 2020, 87, 103423. [Google Scholar] [CrossRef]
- El-Wahed, A.A.A.; Rashwan, E.H.; AlAjmi, M.F.; Khalifa, S.A.M.; Saeed, A.; Zhao, C.; Naggar, Y.A.; Guo, Z.; Musharraf, S.G.; Wang, K. Sidr Honeys Physical and Chemical Characterization, a Comprehensive Approach through LC-MS/MS, NMR, and GC-MS Analysis. Separations 2023, 10, 372. [Google Scholar] [CrossRef]
- Wang, Y.; Suo, Y.; Sun, Y.; You, J. Determination of triterpene acids from 37 different varieties of raspberry using pre-column derivatization and HPLC fluorescence detection. Chromatographia 2016, 79, 1515–1525. [Google Scholar] [CrossRef]
- Tedesco, R.; Barbaro, E.; Zangrando, R.; Rizzoli, A.; Malagnini, V.; Gambaro, A.; Fontana, P.; Capodaglio, G. Carbohydrate determination in honey samples by ion chromatography–mass spectrometry (HPAEC-MS). Anal. Bioanal. Chem. 2020, 412, 5217–5227. [Google Scholar] [CrossRef]
- Duckstein, S.M.; Stintzing, F.C. Investigation on the phenolic constituents in Hamamelis virginiana leaves by HPLC-DAD and LC-MS/MS. Anal. Bioanal. Chem. 2011, 401, 677–688. [Google Scholar] [CrossRef]
- Sundaram, R.S.; Ramanathan, M.; Rajesh, R.; Satheesh, B.; Saravanan, D. LC-MS quantification of rosmarinic acid and ursolic acid in the Ocimum sanctum Linn. leaf extract (Holy basil, Tulsi). J. Liq. Chromatogr. Relat. Technol. 2012, 35, 634–650. [Google Scholar] [CrossRef]
- Keyhanian, S.; Stahl-Biskup, E. Phenolic constituents in dried flowers of Aloe vera (Aloe barbadensis) and their in vitro antioxidative capacity. Planta Med. 2007, 73, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Classification for β-lactamases: Historical perspectives. Expert Rev. Anti. Infect. Ther. 2023, 21, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Sahu, M.K.; Nayak, A.K.; Hailemeskel, B.; Eyupoglu, O.E. Exploring recent updates on molecular docking: Types, method, application, limitation & future prospects. Int. J. Pharm. Res. Allied Sci. 2024, 13, 24–40. [Google Scholar]
- He, Y.; Lei, J.; Pan, X.; Huang, X.; Zhao, Y. The hydrolytic water molecule of Class A β-lactamase relies on the acyl-enzyme intermediate ES* for proper coordination and catalysis. Sci. Rep. 2020, 10, 10205. [Google Scholar] [CrossRef]
- Weljie, A.M.; Newton, J.; Mercier, P.; Carlson, E.; Slupsky, C.M. Targeted Profiling: Quantitative Analysis of 1H NMR Metabolomics Data. Anal. Chem. 2006, 78, 4430–4442. [Google Scholar] [CrossRef]
- Gu, H.; Pan, Z.; Xi, B.; Hainline, B.E.; Shanaiah, N.; Asiago, V.; Gowda, G.A.N.; Raftery, D. 1H NMR metabolomics study of age profiling in children. NMR Biomed. 2009, 22, 826–833. [Google Scholar] [CrossRef]
- Naama, R.T.A. Evaluation of in-vitro inhibitory effect of honey on some microbial isolate. African J. Bacteriol. Res. 2009, 1, 064–067. [Google Scholar]
- Bush, K. Past and present perspectives on β-lactamases. Antimicrob. Agents Chemother. 2018, 62, 10–1128. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]

| Tests | Results | Standards | |||
|---|---|---|---|---|---|
| HO1 | HO2 | HO4 | HO5 | Limitations of Saudi and Gulf Standard Specifications | |
| Physical tests | |||||
| 1. Color | Dark amber | Light amber | Light amber | Dark amber | |
| 2. Odor | Odorless | Odorless | Odorless | Odorless | Free from foreign odors |
| 3. Taste | Sweet | Sweet | Sweet | Sweet | |
| 4. Purity | Pure | Pure | Pure | Pure | Free from impurities |
| Chemical tests | |||||
| 1. Moisture | 15.80 | 15.20 | 17.20 | 18.40 | Calluna and clover honeys: 23% max.; other kinds: 21% max. |
| 2. Reducing sugars | 74.20 | 74.47 | 77.20 | 72.90 | Nectar honey: 60% min. Honey dew honey: 45% min. |
| - Glucose | 34.20 | 31.96 | 38.40 | 35.60 | |
| - Fructose | 40.00 | 42.51 | 38.80 | 37.30 | |
| - Fructose/glucose ratio | 1.17 | 1.33 | 1.01 | 1.05 | |
| 3. Sucrose | 1.50 | 1.03 | 1.5 | 0.50 | 5% max. |
| 4. Hydroxymethylfurfural (HMF) | 53.70 | 21.10 | 15.40 | 69.10 | 80 mg/kg max. |
| 5. Acidity | 14.00 | 6.40 | 12.00 | 32.00 | 50 meq/kg max. |
| 6. Diastase enzyme | -- | -- | 12 | 16.6 | 3 Goth Scal min. |
| Staphylococcus aureus ATCC 25923 | Klebsiella pneumoniae ATCC 700603 | Pseudomonas auruginodse ATCC 90902 | Salmonella typhimurium ATCC 14028 | Escherichia coli ATCC8739 lot 03801105 | Enterococcus faecalis | Enterobacter sp. | |
|---|---|---|---|---|---|---|---|
| HO2 | 49.7 | 11.6 | 85.9 | 85.5 | 38.5 | 47.8 | 0 |
| HO5 | 90.7 | 18.5 | 85.6 | 85.4 | 49.8 | 59.7 | 6.5 |
| HO4 | 58.7 | 0 | 0 | 6.5 | 33.5 | 38.4 | 0 |
| HO1 | 93.3 | 81.1 | 50.5 | 82.9 | 65.0 | 71.1 | 78.3 |
| Cephalosporin (10 ug/mL) | 98.2 | 97.4 | - | 98.4 | 98.2 | - |
| Bacteria | Staphylococcus aureus ATCC 25923 | Klebsiella pneumoniae ATCC 700603 | Pseudomonas auruginose ATCC 90902 | Salmonella typhimurium ATCC 14028 | Escherichia coli 0157 ATCC 700728 | Enterococcus faecalis | Enterobacter sp. |
|---|---|---|---|---|---|---|---|
| HO2 | 25 | - | 1.5625 | 1.5625 | 12.5 | 25 | - |
| HO5 | 1.5625 | 25 | 1.5625 | 1.5625 | 12.5 | 3.125 | - |
| HO4 | 12.5 | - | - | - | 12.5 | 12.5 | - |
| HO1 | 1.5625 | 1.5625 | 12.5 | 1.5625 | 12.5 | 12.5 | 1.5625 |
| Cephalosporin (10 ug/mL) | 1.5625 | 1.5625 | - | 1.5625 | 1.5625 | - | - |
| N. | Compound | Mode | Formula | Class | m/z | Rt | % vol. | References |
|---|---|---|---|---|---|---|---|---|
| 1 | Arabinose | + | C5H10O5 | Sugar | 151.0357 | 0.265 | 0.09% | [17] |
| 2 | 4-Hydroxybenzoic acid | + | C7H6O3 | Phenolic acid | 139.0068 | 0.3 | 0.13% | [18] |
| 3 | Raffinose | − | C18H32O16 | Sugar | 503.1666 | 2.324 | 0.83% | [19] |
| 4 | Sucrose | − | C12H22O11 | Sugar | 341.1098 | 2.36 | 3% | [20] |
| 5 | Maltotetraose | − | C24H42O21 | Sugar | 665.2105 | 2.386 | 0.19% | [21] |
| 6 | Riboflavin | − | C17H20N4O6 | Vitamin | 375.1089 | 2.395 | 0.08% | [22] |
| 7 | Aloe-emodin | + | C15H10O5 | Anthraquinone | 271.0834 | 2.425 | 0.09% | [23] |
| 8 | Epigallocatechin | + | C15H14O7 | Flavonoid | 307.1038 | 2.434 | 0.08% | [24] |
| 9 | Glucose | − | C6H12O6 | Sugar | 179.0560 | 2.455 | 19.81% | [20] |
| 10 | Epigallocatechin gallate | + | C22H18O11 | Flavonoid | 459.1378 | 2.504 | 0.05% | [24] |
| 11 | Lactic acid | − | C3H6O3 | Organic acid | 89.02452 | 2.559 | 9.43% | [22] |
| 12 | Naringenin | + | C15H12O5 | Flavonoid | 273.0837 | 2.62 | 0.28% | [25] |
| 13 | Linoleic acid | + | C18H32O2 | Fatty acid | 281.0724 | 2.684 | 0.62% | [26] |
| 14 | Biotin | + | C10H16N2O3S | Vitamin | 245.0643 | 2.692 | 0.05% | [27] |
| 15 | Corosolic acid | − | C30H48O4 | Triterpenoid | 472.2012 | 3.126 | 0.11% | [28] |
| 16 | Fructose | − | C6H12O6 | Sugar | 179.0557 | 3.135 | 7.99% | [20] |
| 17 | Cinnamic acid | + | C9H8O2 | Phenolic acid | 149.0210 | 4.872 | 0.57% | [25] |
| 18 | Lactose | − | C12H22O11 | Sugar | 341.1156 | 5.16 | 0.72% | [29] |
| 19 | Gallic acid | + | C7H6O5 | Phenolic acid | 171.0996 | 8.107 | 0.38% | [18] |
| 20 | Ferulic acid | + | C10H10O4 | Phenolic acid | 195.0631 | 12.823 | 0.23% | [18] |
| 21 | Gallic acid hexoside | − | C13H16O10 | Phenolic glycoside | 331.2493 | 19.577 | 0.54% | [30] |
| 22 | Syringic acid | + | C9H10O5 | Phenolic acid | 199.0945 | 20.41 | 0.09% | [18] |
| 23 | Rosmarinic acid | − | C18H16O8 | Phenolic acid | 359.1254 | 22.647 | 0.23% | [31] |
| 24 | Chlorogenic acid | − | C16H18O9 | Phenolic acid | 353.2119 | 25.826 | 0.27% | [18] |
| 25 | Saponarin | + | C27H30O15 | Flavonoid | 595.3831 | 29.398 | 0.04% | [32] |
| 26 | Aloin | + | C21H22O9 | Anthraquinone | 419.2789 | 29.823 | 0.04% | [23] |
| 27 | Caffeic acid | + | C9H8O4 | Phenolic acid | 181.9860 | 40.337 | 0.29% | [18] |
| Bush–Jacoby Group Classification | Name | Protein Data Bank (PDB) Code | Bacteria |
|---|---|---|---|
| Group 1: Cephalosporinases | AmpC-type beta-lactamases | 1KVL | Escherichia coli |
| Group 2: Serine beta-lactamases | |||
| 2a: Penicillinases | TEM-1 beta-lactamase | 1FQG | Escherichia coli |
| 2b: Broad-spectrum beta-lactamases | SHV-1 beta-lactamase | 1SHV | Klebsiella pneumoniae |
| 2be: Extended-spectrum beta-lactamases (ESBLs) | CTX-M-14 beta-lactamase | 4HBU | Escherichia coli |
| 2br: Inhibitor-resistant beta-lactamases | TEM-30 beta-lactamase | 1LHY | Escherichia coli |
| 2c: Carbenicillinases | PSE-4 beta-lactamase | 1G68 | Pseudomonas aeruginosa |
| 2d: Cloxacillin-hydrolyzing beta-lactamases (OXA-type) | OXA-10 beta-lactamase | 1FOF | Pseudomonas aeruginosa |
| 2f: Carbapenemases | KPC-2 beta-lactamase | 4ZBE | Klebsiella pneumoniae |
| Group 3: Metallo-beta-lactamases (MBLs) | NDM-1 beta-lactamase | 4EXS | Klebsiella pneumoniae |
| Group 4: Penicillinases that do not fit into other groups | Double mutation, E166D:N170Q, of class A enzyme | 1GHI | Staphylococcus aureus |
| Compound Name | 1KVL | 1FQG | 1SHV | 4HBU | 1LHY | 1G68 | 1FOF | 4ZBE | 4EXS | 1GHI | Total Consensus Score a |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4-Hydroxybenzoic acid | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 2 |
| Aloe-emodin | 3 | 0 | 2 | 2 | 1 | 1 | 0 | 3 | 9 | 0 | 21 |
| Aloin | 1 | 4 | 7 | 7 | 0 | 6 | 4 | 3 | 5 | 2 | 39 |
| Caffeic acid | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 3 |
| Chlorogenic acid | 2 | 6 | 2 | 1 | 9 | 4 | 7 | 2 | 2 | 8 | 43 |
| Cinnamic acid | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 4 |
| Corosolic acid | 3 | 1 | 0 | 3 | 3 | 0 | 0 | 6 | 8 | 3 | 27 |
| Epigallocatechin gallate | 6 | 8 | 0 | 7 | 0 | 0 | 5 | 9 | 4 | 6 | 45 |
| Epigallocatechin | 9 | 0 | 4 | 2 | 6 | 7 | 3 | 2 | 7 | 0 | 40 |
| Ferulic acid | 0 | 0 | 0 | 1 | 0 | 3 | 1 | 1 | 0 | 1 | 7 |
| Gallic acid hexoside | 0 | 4 | 11 | 9 | 7 | 10 | 8 | 1 | 1 | 3 | 54 |
| Gallic acid | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Linoleic acid | 0 | 1 | 1 | 0 | 3 | 0 | 1 | 0 | 1 | 0 | 7 |
| Naringenin | 0 | 0 | 2 | 0 | 3 | 1 | 2 | 1 | 4 | 1 | 14 |
| Rosmarinic acid | 4 | 8 | 10 | 4 | 9 | 5 | 7 | 3 | 1 | 8 | 59 |
| Saponarin | 5 | 9 | 0 | 5 | 0 | 0 | 3 | 9 | 0 | 9 | 40 |
| Syringic acid | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Avibactam | 3 | 4 | 5 | 4 | 5 | 5 | 6 | 6 | 3 | 4 | 45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aati, H.; Lithy, N.M.; Aati, S.Y.; Khanfar, M.A.; Hassan, H.M.; Bahr, H.S. Antimicrobial and Anti-Biofilm Activities of Medicinal Plant-Derived Honey Against ESKAPE Pathogens: Insights into β-Lactamase Inhibition via Metabolomics and Molecular Modeling Studies. Processes 2025, 13, 1294. https://doi.org/10.3390/pr13051294
Aati H, Lithy NM, Aati SY, Khanfar MA, Hassan HM, Bahr HS. Antimicrobial and Anti-Biofilm Activities of Medicinal Plant-Derived Honey Against ESKAPE Pathogens: Insights into β-Lactamase Inhibition via Metabolomics and Molecular Modeling Studies. Processes. 2025; 13(5):1294. https://doi.org/10.3390/pr13051294
Chicago/Turabian StyleAati, Hanan, Nadia M. Lithy, Sultan Y. Aati, Mohammad A. Khanfar, Hossam M. Hassan, and Hebatallah S. Bahr. 2025. "Antimicrobial and Anti-Biofilm Activities of Medicinal Plant-Derived Honey Against ESKAPE Pathogens: Insights into β-Lactamase Inhibition via Metabolomics and Molecular Modeling Studies" Processes 13, no. 5: 1294. https://doi.org/10.3390/pr13051294
APA StyleAati, H., Lithy, N. M., Aati, S. Y., Khanfar, M. A., Hassan, H. M., & Bahr, H. S. (2025). Antimicrobial and Anti-Biofilm Activities of Medicinal Plant-Derived Honey Against ESKAPE Pathogens: Insights into β-Lactamase Inhibition via Metabolomics and Molecular Modeling Studies. Processes, 13(5), 1294. https://doi.org/10.3390/pr13051294








