Metabolic Engineering and Fermentation Process Strategies for L-Tryptophan Production by Escherichia coli
Abstract
1. Introduction
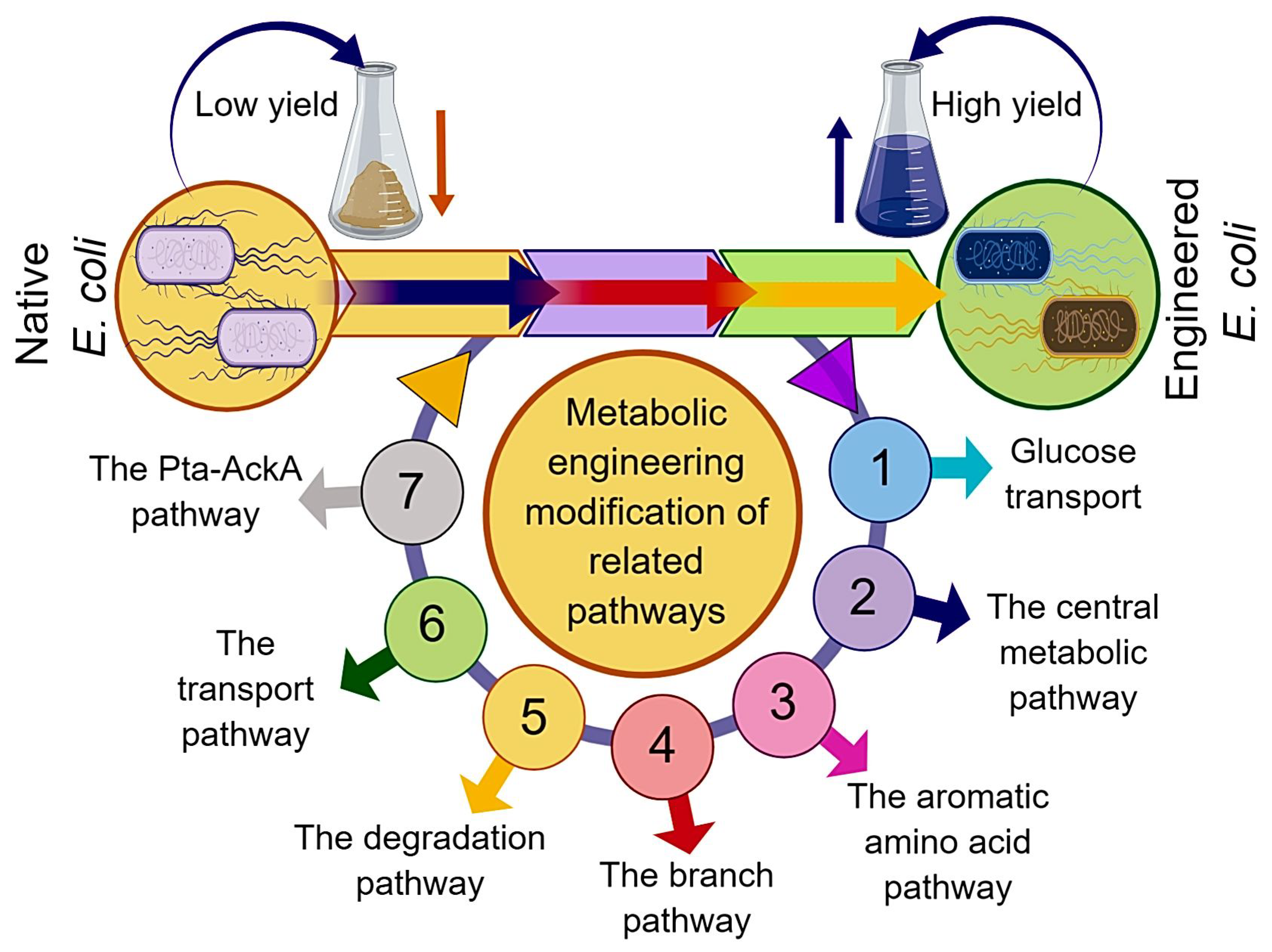
2. Metabolic Engineering Modification of Related Pathways
2.1. Glucose Transport
2.2. The Central Metabolic Pathway
2.3. The Aromatic Amino Acid Common Pathway and the L-tryptophan Branch Pathway
2.4. The Branch Pathway
2.5. The Degradation Pathway
2.6. The Transport Pathway
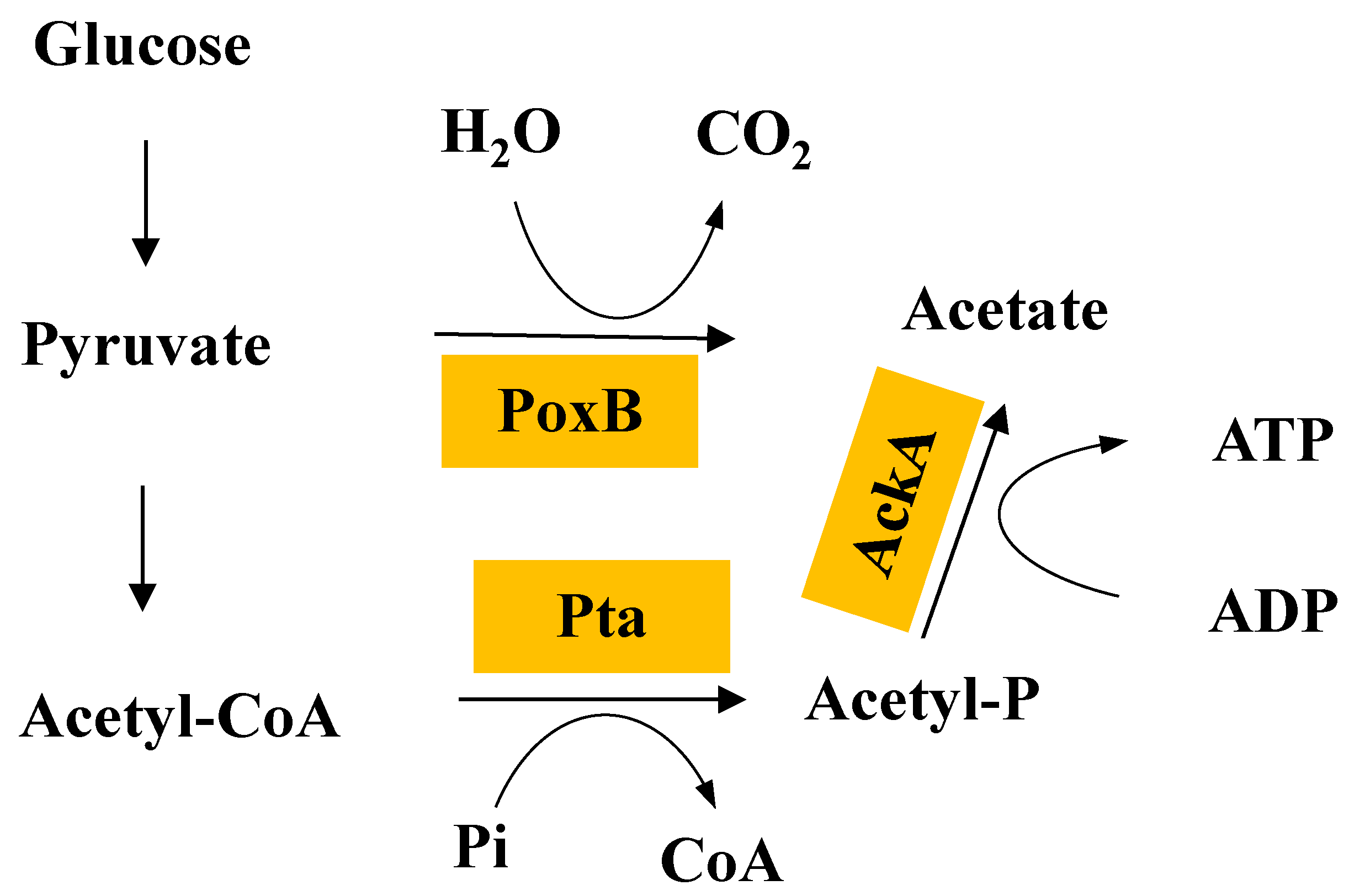
2.7. The Pta-AckA Pathway
2.8. The Polyhydroxybutyrate Synthesis Pathway
3. Transcriptional Regulation by Regulatory Factors
4. Fermentation Process Strategies
4.1. Optimization of Fermentation Conditions
4.2. Addition of Related Substances
5. Conclusions and Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GLC | glucose |
| G6P | glucose-6-phosphate |
| F6P | fructose-6-phosphate |
| F-1,6-BP | fructose-1, 6-bisphosphate |
| GA3P | glyceraldehyde 3-phosphate |
| PG | phosphoglycerate |
| PEP | phosphoenolpyruvate |
| PYR | pyruvate |
| AcCoA | acetyl coenzyme A |
| CisACO | cis-aconitate |
| OAA | oxaloacetate |
| CIT | citric acid |
| ICI | iso-citric acid |
| MAL | malate |
| SUCC | succinate |
| SUCCoA | Succinate coenzyme A |
| 2-OXO | 2-oxoglutarate |
| FUM | fumarate; |
| G-LAC | 6-phospho-glucono-1, 5-lactone; |
| 6-PGT | 6-Phospho-gluconate; |
| Ru5P | ribulose 5-phosphate |
| R5P | ribose 5-phosphate |
| X5P | xylulose 5-phosphate |
| S7P | sedoheptulose 7-phosphate |
| GA3P | glyceraldehyde 3-phosphate |
| E4P | erythrose 4-phosphate; |
| DAHP | 3-deoxy-d-arobino-heptulosonate 7-phosphate; |
| DHS | 3-dehydroshikimate; |
| DHQ | 3-dehydroquinate |
| SHIK | shikimate |
| S3P | shikimate 3-phosphate |
| NADP | oxidized nicotinamide adenine dinucleotide phosphate |
| NADPH | reduced nicotinamide adenine dinucleotide phosphate; |
| ANTA | anthranilate |
| CHA | chorismate |
| I3GP | indole 3-glycerolphosphate |
| Ser | serine |
| Trp | tryptophan |
| Phe | phenylalanine |
| Tyr | tyrosine |
References
- Hermann, T. Industrial production of amino acids by coryneform bacteria. J. Biotechnol. 2003, 104, 155–172. [Google Scholar] [CrossRef]
- Ikeda, M.; Nakagawa, S. The Corynebacterium glutamicum genome: Features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 2003, 62, 99–109. [Google Scholar] [CrossRef]
- Kirchner, O.; Tauch, A. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 2003, 104, 287–299. [Google Scholar] [CrossRef]
- Leuchtenberger, W. Amino Acids—Technical Production and Use. In Biotechnology: Products of Primary Metabolism; Rehm, H.-J., Reed, G., Eds.; Verlagsgesellschaft mbH: Weinheim, Germany, 1996; Chapter 14a; pp. 465–502. [Google Scholar]
- Leuchtenberger, W.; Huthmacher, K.; Drauz, K. Biotechnological production of amino acids and derivatives: Current status and prospects. Appl. Microbiol. Biotechnol. 2005, 69, 1–8. [Google Scholar] [CrossRef]
- Ikeda, M. Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 2006, 69, 615–626. [Google Scholar] [CrossRef]
- Sanchez, S.; Demain, A.L. Microbial primary metabolites: Biosynthesis and perspectives. In Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology; Flickinger, M.C., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; pp. 1–16. [Google Scholar]
- Pontrelli, S.; Chiu, T.Y.; Lan, E.I.; Chen, F.Y.H.; Chang, P.; Liao, J. Escherichia coli as a host for metabolic engineering. Metab. Eng. 2018, 50, 16–46. [Google Scholar] [CrossRef]
- Azuma, S.; Tsunekawa, H.; Okabe, M.; Okamoto, R.; Aiba, S. Hyper-production of L-trytophan via fermentation with crystallization. Appl. Microbiol. Biotechnol. 1993, 39, 471–476. [Google Scholar] [CrossRef]
- Zhao, Z.J.; Zou, C.; Zhu, Y.X.; Dai, J.; Chen, S.; Wu, D.; Wu, J.; Chen, J. Development of L-tryptophan production strains by defined genetic modification in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2011, 38, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Aiba, S.; Tsunekawa, H.; Imanaka, T. New approach to tryptophan production by Escherichia coli: Genetic manipulation of composite plasmids in vitro. Appl. Microbiol. Biotechnol. 1982, 43, 289–297. [Google Scholar]
- Caligiuri, M.G.; Bauerle, R. Subunit Communication in the Anthranilate Synthase Complex from Salmonella typhimurium. Science 1991, 252, 1845–1848. [Google Scholar] [CrossRef]
- Chan, E.C.; Tsai, H.L.; Chen, S.L.; Mou, D.G. Amplification of the tryptophan operon gene in Escherichia coli chromosome to increase L-tryptophan biosynthesis. Appl. Microbiol. Biotechnol. 1993, 40, 301–305. [Google Scholar] [CrossRef]
- De, A.R.; Lara, A.R.; Hernández, V.; Hernández-Montalvo, V.; Gosset, G.; Bolívar, F.; Ramírez, O.T. Replacement of the glucose phosphotransferase transport system by galactose permease reduces acetate accumulation and improves process performance of Escherichia coli for recombinant protein production without impairment of growth rate. Metab. Eng. 2006, 8, 281–290. [Google Scholar]
- Deutscher, J.; Aké, F.M.; Derkaoui, M.; Zébré, A.C.; Cao, T.N.; Bouraoui, H.; Kentache, T.; Mokhtari, A.; Milohanic, E.; Joyet, P. The bacterial phosphoenolpyruvate:Carbohydrate phosphotransferase system: Regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 2014, 78, 231–256. [Google Scholar] [CrossRef] [PubMed]
- Sonja, K.B. Glucose Transport in Escherichia coli Mutant Strains with Defects in Sugar Transport Systems. J. Bacteriol. 2012, 194, 5897–5908. [Google Scholar]
- Chen, Y.; Liu, Y.; Ding, D.; Cong, L.; Zhang, D. Rational design and analysis of an Escherichia coli strain for high-efficiency tryptophan production. J. Ind. Microbiol. Biotechnol. 2018, 45, 1–11. [Google Scholar] [CrossRef]
- Wu, T.; Zhao, J.; Mao, X. Effect of PTS modifications on L-tryptophan production in Escherichia coli. Chin. J. Biotechnol. 2017, 33, 1877–1882. [Google Scholar]
- Gu, P.; Kang, J.; Yang, F.; Wang, Q.; Liang, Q.; Qi, Q. The improved L-tryptophan production in recombinant Escherichia coli by expressing the polyhydroxybutyrate synthesis pathway. Appl. Microbiol. Biotechnol. 2013, 97, 4121–4127. [Google Scholar] [CrossRef]
- Gu, P.; Fan, Y.; Kang, J.; Wang, Q.; Qi, Q. One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of L-tryptophan in Escherichia coli. Microb. Cell. Fact. 2012, 11, 30. [Google Scholar] [CrossRef]
- Snoep, J.L.; Arfman, N.; Yomano, L.P.; Fliege, R.K.; Conway, T.; Ingram, L.O. Reconstruction of glucose uptake and phosphorylation in a glucose-negative mutant of Escherichia coli by using Zymomonas mobilis genes encoding the glucose facilitator protein and glucokinase. J. Bacteriol. 1994, 176, 2133–2135. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, L.; Chen, S.; Wu, J. Phosphoenolpyruvate: Glucose phosphotransferase system modification increases the conversion rate during L-tryptophan production in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2017, 44, 1–11. [Google Scholar] [CrossRef]
- Singh, R.; White, D.; Demirel, Y.; Kelly, R.; Noll, K.; Blum, P. Uncoupling fermentative synthesis of molecular hydrogen from biomass formation in Thermotoga maritima. Appl. Environ. Microbiol. 2018, 84, e00998-18. [Google Scholar] [CrossRef]
- Singh, R.; Tevatia, R.; White, D.; Demirel, Y.; Blum, P. Comparative kinetic modeling of growth and molecular hydrogen overproduction by engineered strains of Thermotoga maritima. Int. J. Hydrog. Energy. 2019, 44, 7125–7136. [Google Scholar] [CrossRef]
- Ongay, R. Tryptophan Biosynthesis by Genetically Engineered Escherichia coli Utilizing Different Carbon Sources; Texas A & M University: College Station, TX, USA, 1994. [Google Scholar]
- Siddiquee, K.A.Z.; Arauzo-Bravo, M.J.; Shimizu, K. Effect of a pyruvate kinase (pykF-gene) knockout mutation on the control of gene expression and metabolic fluxes in Escherichia coli. FEMS Microbiol. Lett. 2004, 235, 25–33. [Google Scholar] [CrossRef]
- Al, Z.S.K.; Arauzobravo, M.J.; Shimizu, K. Metabolic flux analysis of pykF gene knockout Escherichia coli based on 13C-labeling experiments together with measurements of enzyme activities and intracellular metabolite concentrations. Appl. Microbiol. Biotechnol. 2004, 63, 407–417. [Google Scholar]
- Kedar, P.; Colah, R.; Shimizu, K. Proteomic investigation on the gene knockout Escherichia coli for aromatic amino acid production. Enzyme Microb. Tech. 2007, 41, 455–465. [Google Scholar] [CrossRef]
- Niersbach, M.; Kreuzaler, F.; Geerse, R.H.; Postma, W.; Hirsch, H.J. Cloning and nucleotide sequence of the Escherichia coli K-12 ppsA gene, encoding PEP synthase. Mol. Gen. Genet. 1992, 231, 332–336. [Google Scholar] [PubMed]
- Liao, J.C.; Chao, Y.P.; Patnaik, R. Alteration of the biochemical valves in the central metabolism of Escherichia coli. Ann. NY Acad. Sci. 1994, 745, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, T.; Yu, J.L.; Xu, Q.S.; Guo, C.J.; Huang, Y.W. Modification of the central metabolism of E. coli and its effect on tryptophan yield. Chin. Med. Biotechnol. 2008, 3, 93–97. [Google Scholar]
- Tan, Z.; Zhu, X.; Chen, J.; Li, Q.; Zhang, X. Activating phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in combination for improvement of succinate production. Appl. Environ. Microb. 2013, 79, 4838–4844. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Tsujimoto, K.; Kurahashi, O. Mutational analysis of the feedback sites of phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. Appl. Environ. Microb. 1997, 63, 761–762. [Google Scholar]
- Bongaerts, J.; Krämer, M.; Müller, U.; Raeven, L.; Wubbolts, M. Metabolic Engineering for Microbial Production of Aromatic Amino Acids and Derived Compounds. Metab. Eng. 2001, 3, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Jossek, R.; Bongaerts, J.; Sprenger, G.A. Characterization of a new feedback-resistant 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase AroF of Escherichia coli. FEMS Microbiol. Lett. 2001, 202, 145–148. [Google Scholar] [CrossRef]
- Balderashernández, V.E.; Sabidoramos, A.; Silva, P.; Cabreravalladares, N.; Hernándezchávez, G.; Báezviveros, J.L.; Martínez, A.; Bolívar, F.; Gosset, G. Metabolic engineering for improving anthranilate synthesis from glucose in Escherichia coli. Microb. Cell. Fact. 2009, 8, 1–12. [Google Scholar]
- Li, J.X.; Guo, C.J.; Liu, Y.; Lin, W.P.; Zhang, X.M.; Xu, Q.S. Cloning and expression of trpED encoding ANTA synthase Escherichia coli. Lett. Biotech. 2007, 18, 183–185. [Google Scholar]
- Engelberg-Kulka, H.; Amiel, A.; Miller, C.; Schoulaker-Schwarz, R. Studies on the involvement of the UGA readthrough process in the mechanism of attenuation of the tryptophan operon of Escherichia coli. Mol. Gen. Genet. 1982, 188, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Tobey, K.L.; Grant, G.A. The nucleotide sequence of the serA gene of Escherichia coli and the amino acid sequence of the encoded protein, D-3-phosphoglycerate dehydrogenase. J. Biol. Chem. 1986, 261, 12179–12183. [Google Scholar]
- Dodge, T.C.; Gerstner, J.M. Optimization of the glucose feed rate profile for the production of tryptophan from recombinant E coli. J. Chem. Technol. Biot. 2002, 77, 1238–1245. [Google Scholar] [CrossRef]
- Liu, D.X.; Fan, C.S.; Tao, J.H.; Liang, G.X.; Gao, S.E.; Wang, H.J.; Li, X.; Song, D.X. Integration of E.coli aroG-pheA tandem genes into Corynebacterium glutamicum tyrA locus and its effect on L-phenylalanine biosynthesis. World J. Gastroenterol. 2004, 10, 3683–3687. [Google Scholar] [CrossRef]
- Li, G.; Young, K.D. A new suite of tnaA mutants suggests that Escherichia coli tryptophanase is regulated by intracellular sequestration and by occlusion of its active site. BMC Microbiol. 2015, 15, 1–17. [Google Scholar] [CrossRef]
- Marínsanguino, A.; Torres, N.V. Optimization of tryptophan production in bacteria. Design of a strategy for genetic manipulation of the tryptophan operon for tryptophan flux maximization. Biotechnol. Progr. 2000, 16, 133–145. [Google Scholar] [CrossRef]
- Zhao, Z.J.; Chen, S.; Wu, D.; Wu, J.; Chen, J. Effect of gene knockouts of L-tryptophan uptake system on the production of L-tryptophan in Escherichia coli. Process Biochem. 2012, 47, 340–344. [Google Scholar] [CrossRef]
- Gu, P.; Fan, Y.; Li, F.; Liang, Q.; Qi, Q. Knocking out analysis of tryptophan permeases in Escherichia coli for improving L-tryptophan production. Appl. Microbiol. Biotechnol. 2013, 97, 6677–6683. [Google Scholar] [CrossRef]
- Liu, R.; Liang, L.; Chen, K.; Ma, J.; Jiang, M.; Wei, P.; Ouyang, P. Fermentation of xylose to succinate by enhancement of ATP supply in metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2012, 94, 959–968. [Google Scholar] [CrossRef]
- Jing, L.I.; Shi, B.C.; Wang, C.Y.; Zhao, Z.J.; Shi, J.P. Effect of engineering of L-tryptophan transport system on L-tryptophan production in Escherichia coli. Sci. Technol. Food Indus. 2017, 15. [Google Scholar]
- Wang, J.; Cheng, L.K.; Wang, J.; Liu, Q.; Shen, T.; Chen, N. Genetic engineering of Escherichia coli to enhance production of L-tryptophan. Appl. Microbiol. Biotechnol. 2013, 97, 7587–7596. [Google Scholar] [CrossRef]
- Liu, L.; Duan, X.; Wu, J. L-Tryptophan Production in Escherichia coli Improved by Weakening the Pta-AckA Pathway. PLoS ONE 2016. [Google Scholar] [CrossRef]
- Suárez, D.C.; Kilikian, B.V. Acetic acid accumulation in aerobic growth of recombinant Escherichia coli. Process Biochem. 2000, 35, 1051–1055. [Google Scholar] [CrossRef]
- Phue, J.N.; Sang, J.L.; Kaufman, J.B.; Negrete, A.; Shiloach, J. Acetate accumulation through alternative metabolic pathways in ackA. Biotechnol. Lett. 2010, 32, 1897–1903. [Google Scholar] [CrossRef]
- Mai, L.; Shanjing, Y.; Kazuyuki, S. Effect of poxB gene knockout on metabolism in Escherichia coli based on growth characteristics and enzyme activities. World J. Microb. Biot. 2007, 23, 573–580. [Google Scholar]
- Kakuda, H.; Hosono, K.; Shiroishi, K.; Ichihara, S. Identification and characterization of the ackA (acetate kinase A)-pta (phosphotransacetylase) operon and complementation analysis of acetate utilization by an ackA-pta deletion mutant of Escherichia coli. J. Biochem. 1994, 116, 916–922. [Google Scholar] [CrossRef]
- Zhao, C.; Cheng, L.K.; Wang, J.; Shen, Z.; Chen, N. Impact of deletion of the genes encoding acetate kinase on production of L-tryptophan by Escherichia coli. Ann. Microbiol. 2016, 66, 1–9. [Google Scholar] [CrossRef]
- Klein, A.H.; Shulla, A.; Reimann, S.A.; Keating, D.H.; Wolfe, A.J. The Intracellular Concentration of Acetyl Phosphate in Escherichia coli Is Sufficient for Direct Phosphorylation of Two-Component Response Regulators. J. Bacteriol. 2007, 189, 5574–5581. [Google Scholar] [CrossRef]
- Chang, D.E.; Shin, S.; Rhee, J.S.; Pan, J.G. Acetate metabolism in a pta mutant of Escherichia coli W3110: Importance of maintaining acetyl coenzyme A flux for growth and survival. J. Bacteriol. 1999, 181, 6656–6663. [Google Scholar] [PubMed]
- Mccleary, W.R.; Stock, J.B. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 1994, 269, 31567–31572. [Google Scholar]
- Tyo, K.E.; Fischer, C.R.; Simeon, F.; Stephanopoulos, G. Analysis of polyhydroxybutyrate flux limitations by systematic genetic and metabolic perturbations. Metab. Eng. 2010, 12, 187–195. [Google Scholar] [CrossRef]
- Gottesman, S. Bacterial regulation: Global regulatory networks. Annu. Rev. Genet. 1984, 18, 415–441. [Google Scholar] [CrossRef] [PubMed]
- Schmidm, J.W.; Mauch, K.; Reuss, M.; Gilles, E.D.; Kremling, A. Metabolic design based on a coupled gene expression-metabolic network model of tryptophan production in Escherichia coli. Metab. Eng. 2004, 6, 364–377. [Google Scholar] [CrossRef]
- Ramseier, T.M.; Bledig, S.; Michotey, V.; Feghali, R.; Saier, M.H.J. The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol. Microbiol. 1995, 16, 1157–1169. [Google Scholar] [CrossRef]
- Ow, S.W.; Lee, M.Y.; Nissom, P.M.; Philp, R.; Oh, K.W.; Yap, G.S. Inactivating FruR global regulator in plasmid-bearing Escherichia coli alters metabolic gene expression and improves growth rate. J. Biotechnol. 2007, 131, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Kurata, H.; Shimizu, K. Effect of cra gene mutation on the metabolism of Escherichia coli for a mixture of multiple carbon sources. Adv. Biosci. Biotechnol. 2013, 4, 477–486. [Google Scholar] [CrossRef]
- Liu, L.; Duan, X.; Wu, J. Modulating the direction of carbon flow in Escherichia coli to improve L-tryptophan production by inactivating the global regulator FruR. J. Biotechnol. 2016, 231, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Sabnis, N.A.; Yang, H.; Romeo, T. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 1995, 270, 29096–29104. [Google Scholar] [CrossRef]
- Lijun, L.U.; Bin, Z.; Hong, Z.; Xinyao, L.U.; Fang, H.; Jian, S. Improved production of L-tryptophan in Escherichia coli with csrB and tktA overexpression. Chin. J. Appl. Environ. Biol. 2015, 21, 647–651. [Google Scholar]
- Wang, J.G.; Fan, C.S.; Wu, Y.Q.; Jin, R.L.; Liu, D.X.; Shang, L.; Jiang, P.H. Regulation of aroP expression by tyrR gene in Escherichia coli. Acta Bioch. Bioph. Sin. 2003, 35, 993–997. [Google Scholar]
- Shang, L.; Fan, C.S.; Jin, R.L.; Liu, D.X.; Wang, J.G.; Yin, J.; Song, D.X. Knockout of tyrR gene in Escherichia coli and its effects on the phenylalanine biosynthesis. Acta Bioch. Et. Bioph. Sin. 2003, 35, 728–733. [Google Scholar]
- Tripet, B.P.; Goel, A.; Copie, V. Internal dynamics of the tryptophan repressor (TrpR) and two functionally distinct TrpR variants, L75F-TrpR and A77V-TrpR, in their L-Trp-bound forms. Biochemistry 2011, 50, 5140–5153. [Google Scholar] [CrossRef]
- Henkin, T.M.; Yanofsky, C. Regulation by transcription attenuation in bacteria: How RNA provides instructions for transcription termination/antitermination decisions. Bioessays 2002, 24, 700–707. [Google Scholar] [CrossRef]
- Niu, H.; Li, R.; Liang, Q.; Qi, Q.; Li, Q.; Gu, P. Metabolic engineering for improving l-tryptophan production in Escherichia coli. J. Indus. Microbiol. Biotechnol. 2019, 46, 55–65. [Google Scholar] [CrossRef]
- Cheng, L.K.; Wang, J.; Xu, Q.Y.; Xie, X.X.; Zhang, Y.J.; Zhao, C.G.; Chen, N. Effect of feeding strategy on L-tryptophan production by recombinant Escherichia coli. Ann. Microbiol. 2012, 62, 1625–1634. [Google Scholar] [CrossRef]
- Luo, W.; Huang, J.; Zhu, X.C.; Huang, L.; Cai, J.; Xu, Z.N. Enhanced Production of L-Tryptophan with Glucose Feeding and Surfactant Addition and Related Metabolic Flux Redistribution in the Recombinant Escherichia coli. Food Sci. Biotechnol. 2013, 22, 207–214. [Google Scholar] [CrossRef]
- Cheng, L.K.; Huang, J.; Qin, Y.F.; Xu, Q.Y.; Xie, X.X.; Wen, T.Y.; Chen, N. Effect of the byproduct-acetic acid on L-tryptophan fermentation. Microbiol. Chin. 2010, 37, 166–173. [Google Scholar]
- Zou, C.; Chen, S.; Zhao, Z.J.; Lei, W.; Feng, L.; Wu, D.; Jian, C.; Wu, J. Optimization of L-tryptophan fermentation conditions from recombinant Escherichia coli. Ind. Microbiol. 2012, 42, 25–29. [Google Scholar]
- Cheng, L.K.; Wang, J.; Xu, Q.Y.; Zhao, C.G.; Shen, Z.Q.; Xie, X.X.; Chen, N. Strategy for pH control and pH feedback-controlled substrate feeding for high-level production of L-tryptophan by Escherichia coli. World J. Microbiol. Biotechnol. 2013, 29, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.G.; Xie, X.X.; Cheng, L.K.; Qing-Yang, X.U. Effect of Temperature on the Process of Escherichia coli L-Tryptophan Fermentation. Lett. Biotechnol. 2009, 12, A88. [Google Scholar]
- Chen, L.; Zeng, A.P. Rational design and metabolic analysis of Escherichia coli for effective production of L-tryptophan at high concentration. Appl. Microbiol. Biotechnol. 2017, 101, 59–568. [Google Scholar] [CrossRef] [PubMed]
- Sadeghiyan-Rizi, T.; Fooladi, J.; Sadrai, S. Preliminary Study on Cost-Effective L-Tryptophan Production from Indole and L-Serine by E. Coli Cells. Avicenna. J. Med. Biotechnol. 2016, 8, 188–192. [Google Scholar]
- Lu, J.; Tang, J.; Liu, Y.; Zhu, X.; Zhang, T.; Zhang, X. Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl. Microbiol. Biotechnol. 2012, 93, 2455–2462. [Google Scholar] [CrossRef] [PubMed]
- Perrenoud, A.; Sauer, U. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J. Bacteriol. 2005, 187, 3171–3179. [Google Scholar] [CrossRef]
- Prüss, B.M.; Campbell, J.W.; Van, T.D.; Zhu, C.; Kogan, Y.; Matsumura, P. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 2003, 185, 534–543. [Google Scholar] [CrossRef]


 ) are generally upregulated (red boxes) in P+ fruR cells vs. P+ WT cells. At least two biological samples, each with three technical replicates, were analyzed. Reprinted from Ow et al. [62], with permission from Elsevier. Copyright (2007) Elsevier B.V.
) are generally upregulated (red boxes) in P+ fruR cells vs. P+ WT cells. At least two biological samples, each with three technical replicates, were analyzed. Reprinted from Ow et al. [62], with permission from Elsevier. Copyright (2007) Elsevier B.V.
 ) are generally upregulated (red boxes) in P+ fruR cells vs. P+ WT cells. At least two biological samples, each with three technical replicates, were analyzed. Reprinted from Ow et al. [62], with permission from Elsevier. Copyright (2007) Elsevier B.V.
) are generally upregulated (red boxes) in P+ fruR cells vs. P+ WT cells. At least two biological samples, each with three technical replicates, were analyzed. Reprinted from Ow et al. [62], with permission from Elsevier. Copyright (2007) Elsevier B.V.
| Strains | Gene Modificationa | Optimization Strategies | Fermentative Scale (L) | L-Tryptophan Yield (g·L−1) | Conversion Rate (g·g−1) | Productivity (g·L−1 h−1) | Acetate Content (g·L−1) | Reference |
|---|---|---|---|---|---|---|---|---|
| KW023 | A: trpEDCBA, aroG fbr, galP, galk | --- | 5 | 39.7 | 0.167 | 1.6 | 0.5 | [17] |
| B: pykF, ptsH, trpR, attenuator | ||||||||
| C: downregulation of pta | ||||||||
| S028 | A: trpEDCBA | --- | 1.5 | 40 | 0.15 | 0.6 | --- | [78] |
| B: aroF, aroG, mtr, tnaA, tnaB | ||||||||
| C: ΔaroH:: Ptac-aroGfbr-serA | ||||||||
| ΔpykF-ptsHN12S | A: aroFfbr, trpEfbrD | --- | 3 | 26 | 0.178 | --- | 0.6 | [22] |
| B: trpR, tnaA, pheA, tyrA, pykF | ||||||||
| C: Δpta1:: pta1, ΔptsH:: ptsHN12S | ||||||||
| FB-04(pta1) | A: aroFfbr, trpEfbrD | --- | 3 | 44 | 0.129 | 0.82 | 2.1 | [49] |
| B: trpR, tnaA, pheA, tyrA, | ||||||||
| C: Δpta:: pta1, | ||||||||
| ATCC 11303 | Iranian cane molasses | 10 | 0.43 | --- | --- | --- | [79] | |
| concentration | ||||||||
| Triton X-100 concentration | ||||||||
| Dpta/mtr-Y | A: aroGfbr, trpEfbrDCBA, serA, tktA, ppsA, yddG | --- | 30 | 48.7 | 0.219 | --- | 0.95 | [19] |
| B: trpR, tnaA, pta, tyrA, mtr | ||||||||
| GPT1017 | A: aroGfbr, trpEfbr, tktA | --- | 5 | 16.3 | --- | 0.25 | 1.02 | [45] |
| B: trpR, tnaA, tnaB, ptsH, tyrA, aroP, mtr attenuator | ||||||||
| C: Δtrp promoter:5CPtacs promoter cluster | ||||||||
| W3110-ZDrr | A: trpEDCBA | Glucose feeding rate | 10 | 35.5 | --- | --- | --- | [73] |
| B: trpR, tnaA | PL61 concentration | |||||||
| TRJH/ΔPB | A: trpEDCBA | pH | 30 | 43.7 | --- | --- | 1.15 | [76] |
| B: trpR, tnaA, pta | Glucose Feeding rate | |||||||
| FB-04/pSV-04 | --- | Specific growth rate | 3 | 53.4 | --- | --- | 0.3 | [75] |
| Salt ion concentration | ||||||||
| TRJH | A: trpEDCBA | Glucose feeding rate | 30 | 38.8 | 0.199 | --- | 0.9 | [72] |
| FB-04/pSV03 | A: aroF, trpED | --- | 3 | 14.7 | 0.12 | 0.05 | --- | [44] |
| B: trpR, tnaA, mtr, pheA | ||||||||
| TRTH-Y | A: trpEDCBA, yddG | --- | 30 | 36.3 | --- | 1.01 | --- | [16] |
| B: tnaA | ||||||||
| TRTH/pSV-709 | A: trpEDCBA | Dissolved oxygen level | 5 | 33.9 | --- | --- | 0.9 | [74] |
| Initial glucose concentration | ||||||||
| Glucose feeding rate | ||||||||
| Specific growth rate | ||||||||
| TRTH/pSV-709 | A: trpEDCBA | Temperature | 5 | 30.2 | --- | --- | --- | [77] |
| JB102 | A: aroGfbr, trpEfbr, serA | Glucose feeding rate | 14 | 42.3 | 0.176 | --- | --- | [40] |
| AGX1757/pSC101 | A: trpEfbrD | ANTA concentration | 11 | 54.5 | 0.23 | 0.683 | --- | [9] |
| B: trpR, tnaA | ||||||||
| EMS4-C25/pTC701 | A: trpEDCBA | --- | Shake flask | 9.2 | 0.13 | --- | [13] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Bilal, M.; Luo, H.; Zhao, Y.; Iqbal, H.M.N. Metabolic Engineering and Fermentation Process Strategies for L-Tryptophan Production by Escherichia coli. Processes 2019, 7, 213. https://doi.org/10.3390/pr7040213
Liu L, Bilal M, Luo H, Zhao Y, Iqbal HMN. Metabolic Engineering and Fermentation Process Strategies for L-Tryptophan Production by Escherichia coli. Processes. 2019; 7(4):213. https://doi.org/10.3390/pr7040213
Chicago/Turabian StyleLiu, Lina, Muhammad Bilal, Hongzhen Luo, Yuping Zhao, and Hafiz M. N. Iqbal. 2019. "Metabolic Engineering and Fermentation Process Strategies for L-Tryptophan Production by Escherichia coli" Processes 7, no. 4: 213. https://doi.org/10.3390/pr7040213
APA StyleLiu, L., Bilal, M., Luo, H., Zhao, Y., & Iqbal, H. M. N. (2019). Metabolic Engineering and Fermentation Process Strategies for L-Tryptophan Production by Escherichia coli. Processes, 7(4), 213. https://doi.org/10.3390/pr7040213






