Samnamul (Shoots of Aruncus dioicus) Inhibit Adipogenesis by Downregulating Adipocyte-Specific Transcription Factors in 3T3-L1 Adipocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Sample
2.2. Cell Culture and Differentiation
2.3. Cytotoxicity
2.4. Oil Red O Staining
2.5. Triglyceride (TG) Assay
2.6. Measurement of Total Cholesterol
2.7. Western Blot
2.8. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.9. Measurement of Antioxidant Activity
2.10. Total Phenol and Flavonoid Contents
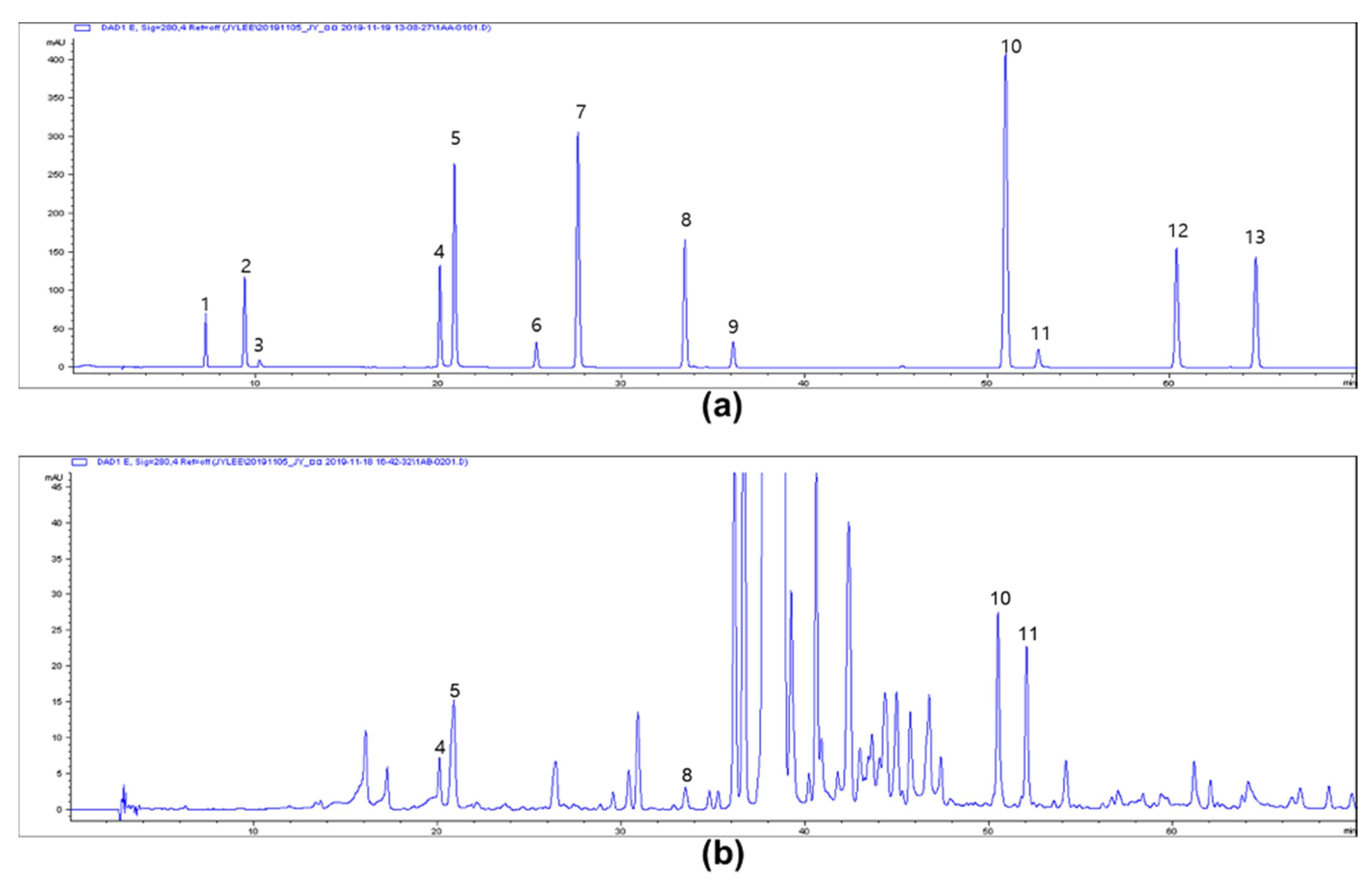
2.11. HPLC Analysis for Phenolics
2.12. Statistics
3. Results
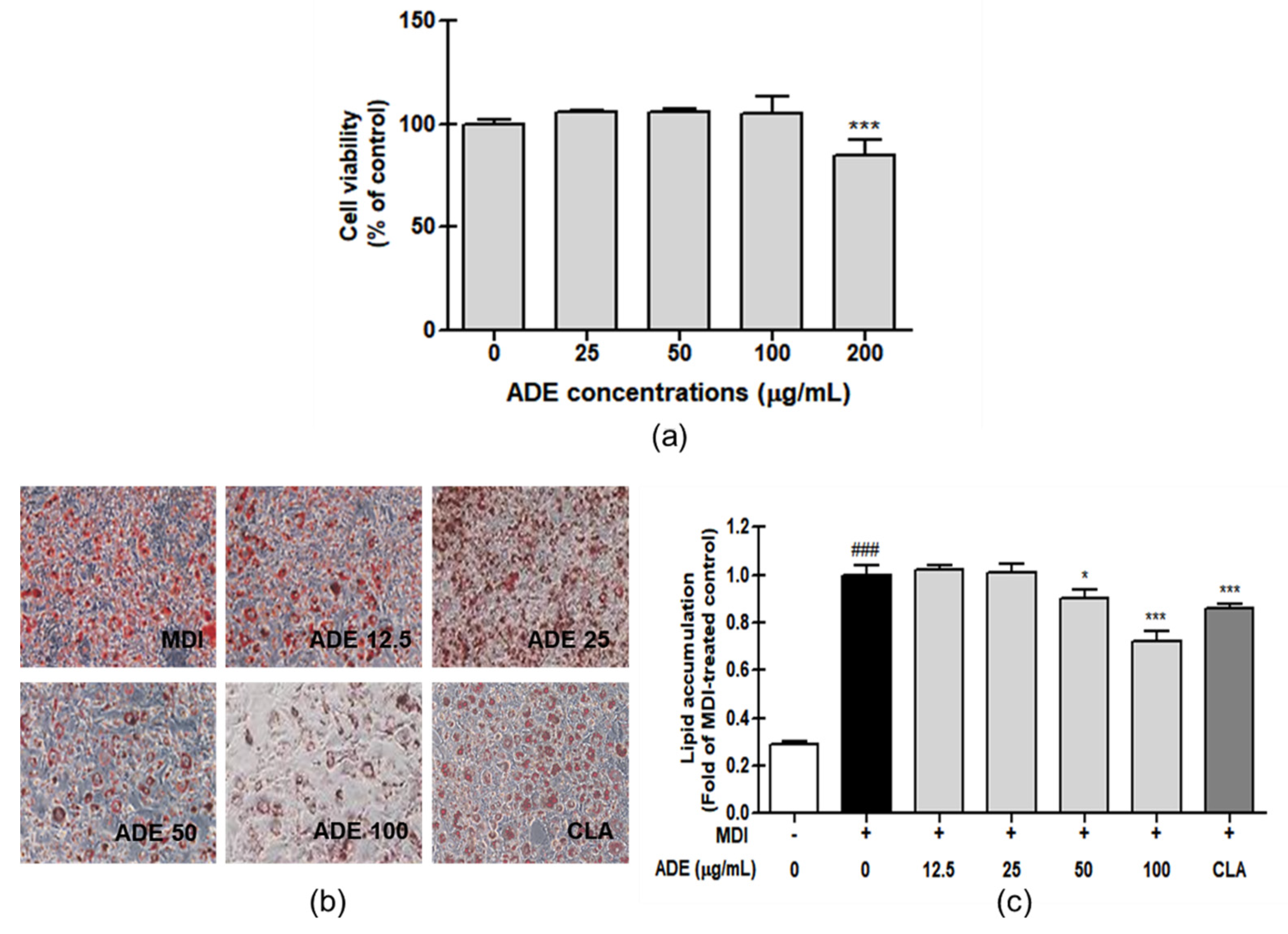
3.1. ADE Has Anti-Adipogenesis Effect on 3T3-L1 Adipocyte
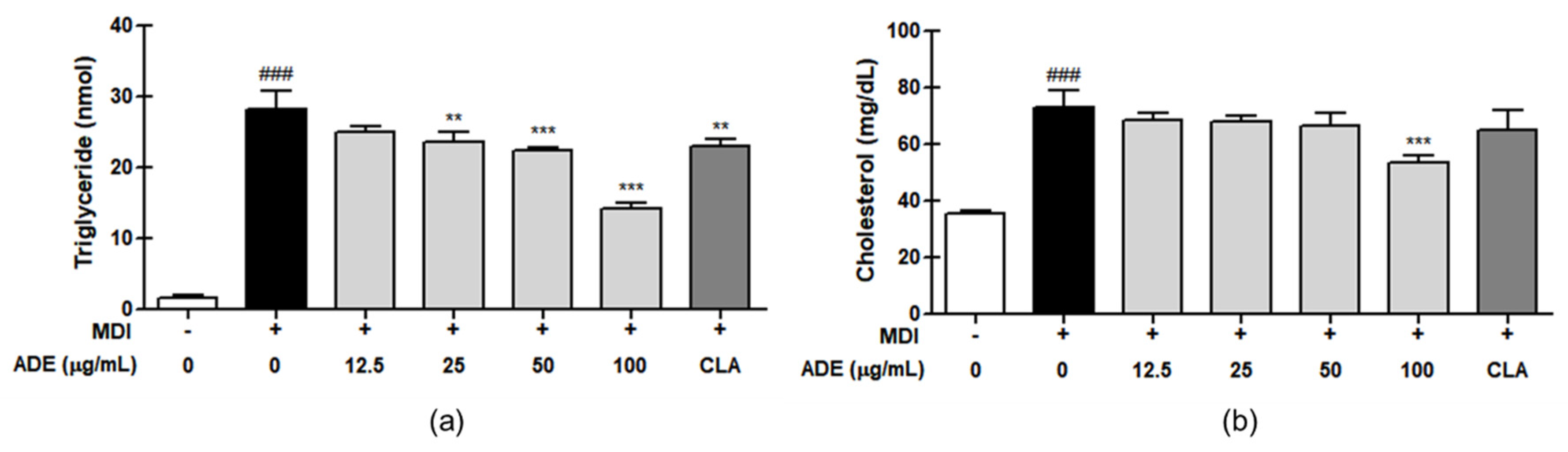
3.2. ADE Reduces the TG and Total Cholesterol Levels in 3T3-L1 Adipocytes
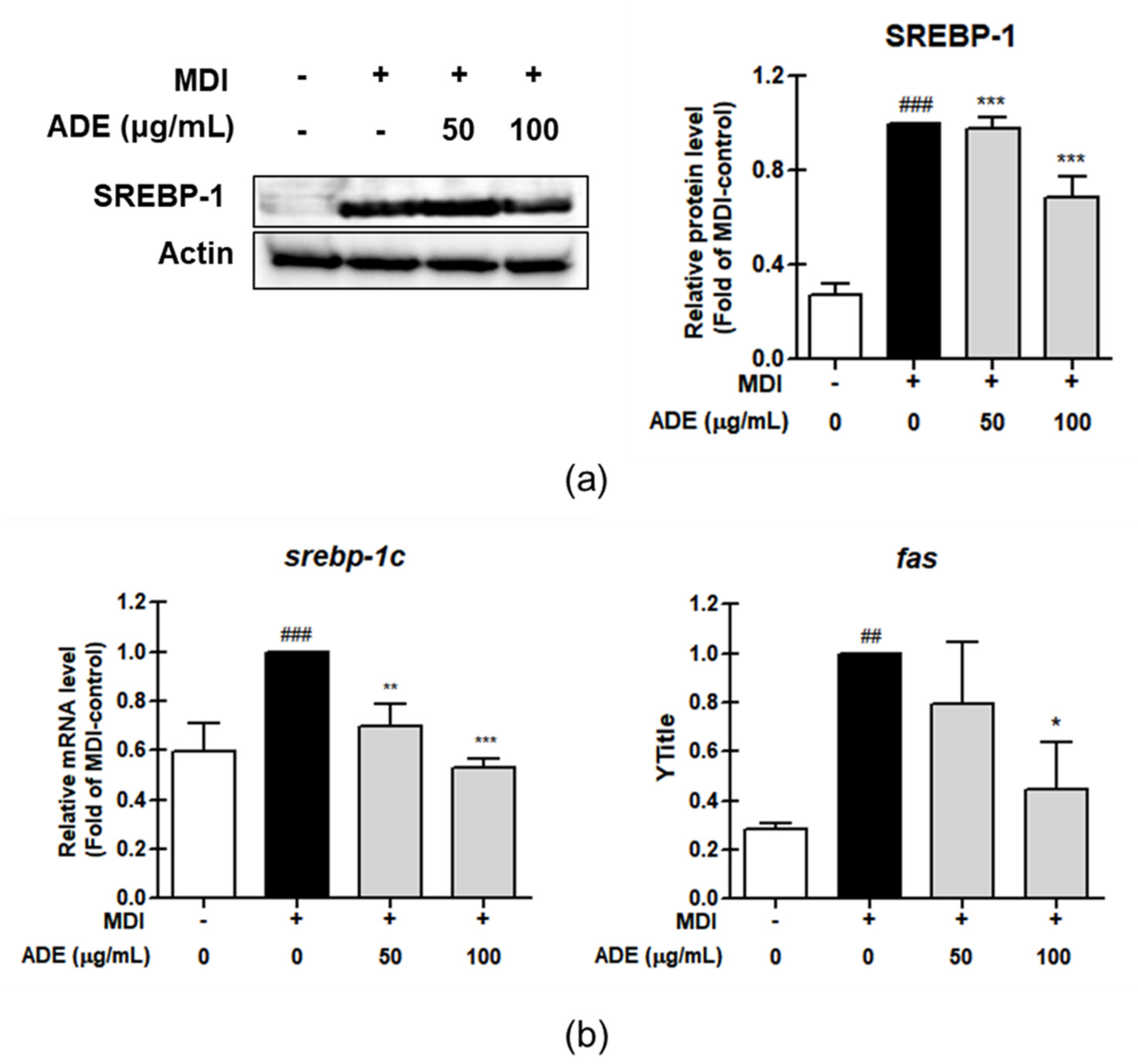
3.3. ADE Induced Anti-Adipogenesis by Controlling Adipogenesis Marker Proteins and mRNA Expression on 3T3-L1 Adipocytes
3.4. ADE Downregulates Lipogenesis Protein and mRNA Expression on 3T3-L1 Adipocytes
3.5. Antioxidant Activities of ADE
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Achike, F.I.; To, N.H.P.; Wang, H.; Kwan, C.Y. Obesity, metabolic syndrome, adipocytes and vascular function: A holistic viewpoint. Clin. Exp. Pharmacol. 2011, 38, 1–10. [Google Scholar] [CrossRef]
- Low, S.; Chin, M.C.; Deurenberg-Yap, M. Review on epidemic of obesity. Ann. Acad. Med. Singap. 2009, 38, 57. [Google Scholar]
- Morshed, A.B.; Kasman, M.; Heuberger, B.; Hammond, R.A.; Hovmand, P.S. A systematic review of system dynamics and agent-based obesity models: Evaluating obesity as part of the global syndemic. Obes. Rev. 2019, 20, 161–178. [Google Scholar] [CrossRef] [Green Version]
- Rashad, I.; Grossman, M. The Economics of Obesity; Public Interest: Denville, NJ, USA, 2004; pp. 104–113. [Google Scholar]
- Ilze, V.; Alvaro, M.V.; Josias, H.H. Natural products in anti-obesity therapy. Nat. Prod. Rep. 2011, 28, 1493–1533. [Google Scholar]
- Mandrup, S.; Lane, M.D. Regulating adipogenesis. J. Biol. 1997, 272, 5367–5370. [Google Scholar] [CrossRef] [Green Version]
- Kersten, S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep. 2001, 2, 282–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turker, I.; Zhang, Y.; Zhang, Y.; Rehman, J. Oxidative stress as a regulator of adipogenesis. FASEB J. 2007, 21, A1053. [Google Scholar]
- Youn, J.S.; Shin, S.Y.; Wu, Y.X.; Hwang, J.Y.; Cho, J.H.; Ha, Y.G.; Kim, J.K.; Park, M.J.; Lee, S.H.; Kim, T.H. Antioxidant and anti-wrinkling effects of Aruncus dioicus var. kamtschaticus extract. Korean J. Food Preserv. 2012, 19, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Kim, A.N.; Lee, K.Y.; Ha, Y.G.; Heo, H.J.; Cho, S.G. Effect of freeze, hot-air, and vacuum drying on antioxidant properties and quality characteristics of samnamul (Aruncus dioicus var. kamtschaticus). Korean J. Food Preserv. 2018, 25, 811–818. [Google Scholar] [CrossRef]
- Zhang, Q.; Kim, H.Y. DNA damage protection and anti-inflammatory activity of different solvent fractions from Aruncus dioicus var. kamtschaticus. Korean J. Plant Res. 2014, 27, 714–719. [Google Scholar] [CrossRef]
- Lee, K.C.; Han, S.K.; Kwon, Y.H.; Jeon, S.R.; Lee, C.W.; Seo, D.J.; Park, W.G. Effects of Shading Treatments on Growth and Physiological Characteristics of Aruncus dioicus var. kamtschaticus (Maxim.) H. Hara Seedling. Korean J. Med. Crop Sci. 2019, 27, 30–37. [Google Scholar] [CrossRef]
- Ahn, H.; Kim, J.; Kim, J.; Auh, J.; Choe, E. In vitro α-glucosidase and pancreatic lipase inhibitory activities and antioxidants of Samnamul (Aruncus dioicus) during rehydration and cooking. Food Sci. Biotechnol. 2014, 23, 1287–1293. [Google Scholar] [CrossRef]
- Kim, J.I.; Yun, J.A.; Jeong, Y.K.; Baek, H.J. Hypoglycemic and hypolipidemic effects of samnamul (Shoot of Aruncus dioicus var. kamtschaticus Hara) in mice fed a high-fat/high-sucrose diet. Food Sci. Biotechnol. 2018, 27, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Park, J.Y.; Kim, H.D.; Lee, S.E.; Lee, J.H.; Lee, Y.; Seo, K.H. Anti-oxidant and anti-adipocyte differentiation of Aster glehni and Aster yomena. J. Nutr. Health 2019, 52, 250–257. [Google Scholar] [CrossRef]
- Ramirez-Zacarias, J.; Castro-Munozledo, F.; Kuri-Harcuch, W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry 1992, 97, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Yang, P.C. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429. [Google Scholar] [PubMed]
- Mishra, K.; Ojha, H.; Chaudhury, N.K. Estimation of antiradical properties of antioxidants using DPPH assay: A critical review and results. Food Chem. 2012, 130, 1036–1043. [Google Scholar] [CrossRef]
- Miller, N.J.; Rice-Evans, C.A. Factors influencing the antioxidant activity determined by the ABTS•+ radical cation assay. Free Radical Res. 1997, 26, 195–199. [Google Scholar] [CrossRef]
- Azlim Almey, A.; Ahmed Jalal Khan, C.; Syed Zahir, I.; Mustapha Suleiman, K.; Aisyah, M.; Kamarul Rahim, K. Total phenolic content and primary antioxidant activity of methanolic and ethanolic extracts of aromatic plants’ leaves. Int. Food Res. 2010, 17, 1077–1084. [Google Scholar]
- Pękal, A.; Pyrzynska, K. Evaluation of aluminium complexation reaction for flavonoid content assay. Food Anal. Methods 2014, 7, 1776–1782. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.Y.; Jang, G.Y.; Lee, S.H.; Kim, K.M.; Lee, J.; Jeong, H.S. Preparation of black soybean (Glycine max L) extract with enhanced levels of phenolic compound and estrogenic activity using high hydrostatic pressure and pre-germination. High. Press. Res. 2018, 38, 177–192. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, J.Y.; Kim, D.H.; Kim, H.D.; Ji, Y.J.; Seo, K.H. Erigeron annuus protects PC12 neuronal cells from oxidative stress induced by ROS-mediated apoptosis. Evid.-Based Compl. Alt. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cristancho, A.G.; Lazar, M.A. Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. 2011, 12, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Shimano, H.; Yahagi, N.; Amemiya Kudo, M.; Hasty, A.H.; Osuga, J.I.; Tamura, Y.; Shionoiri, F.; Iizuka, Y.; Ohashi, K.; Harada, K. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J. Biol. Chem. 1999, 274, 35832–35839. [Google Scholar] [CrossRef] [Green Version]
- Shimano, H. Sterol regulatory element-binding proteins (SREBPs): Transcriptional regulators of lipid synthetic genes. Prog. Lipid Res. 2001, 40, 439–452. [Google Scholar] [CrossRef]
- Gummersbach, C.; Hemmrich, K.; Kröncke, K.-D.; Suschek, C.V.; Fehsel, K.; Pallua, N. New aspects of adipogenesis: Radicals and oxidative stress. Differentiation 2009, 77, 115–120. [Google Scholar] [CrossRef]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radical Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Kim, S.; Ahn, S.H.; Park, J.H.; Park, C.H.; Sin, Y.S.; Shin, G.W.; Kwon, J. Anti-adipogenic effects of viscothionin in 3T3-L1 adipocytes and high fat diet induced obesity mice. Appl. Biol. Chem. 2020, 63, 1–9. [Google Scholar] [CrossRef]
- Ali, A.T.; Hochfeld, W.E.; Myburgh, R.; Pepper, M.S. Adipocyte and adipogenesis. Eur. J. Cell Biol. 2013, 92, 229–236. [Google Scholar] [CrossRef]
- Sarjeant, K.; Stephens, J.M. Adipogenesis. Csh. Perspect. Biol. 2012, 4, a008417. [Google Scholar] [CrossRef] [Green Version]
- Magalhães, L.M.; Segundo, M.A.; Reis, S.; Lima, J.L. Methodological aspects about in vitro evaluation of antioxidant properties. Anal. Chim. Acta. 2008, 613, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Bae, J.H.; Jung, H.Y.; Kim, J.H.; Park, D.S. Antioxidant activity in water and methanol extracts from Korean edible wild plants. J. Korean Soc. Food Sci. Nutr. 2011, 40, 29–36. [Google Scholar] [CrossRef]
- Mashmoul, M.; Azlan, A.; Khaza’ai, H.; Yusof, B.N.M.; Noor, S.M. Saffron: A natural potent antioxidant as a promising anti-obesity drug. Antioxidants 2013, 2, 293–308. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.S.; Kwon, Y.; Choe, S.Y.; Hong, S.M.; Yoo, H.; Goto, T.; Kawada, T.; Choi, H.S.; Joe, Y.; Chung, H.T. Quercetin reduces obesity-induced hepatosteatosis by enhancing mitochondrial oxidative metabolism via heme oxygenase-1. Nutr. Metab. 2015, 12, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvajal-Aldaz, D.G. Inhibition of Adipocyte Differentiation in 3T3-L1 Cell Line by Quercetin or Isorhamnetin. Master’s Theses, Louisiana State University, Baton Rouge, LA, USA, December 2012. [Google Scholar]

| Gene 1 | Sequence (5′→3′) | |
|---|---|---|
| adipoq | Forward | AATTCCCAGGAAGATGAAGG |
| Reverse | GAAGAACAAGCCAAAGACCA | |
| ap2 | Forward | GGAAGCTTGTCTCCAGTGAA |
| Reverse | CGCCTTTCATAACACATTCC | |
| srebp-1c | Forward | TTGAGGATAGCCAGGTCAAA |
| Reverse | ACAAAGAGAAGAGCCAAGCA | |
| fas | Forward | AAGCAAGAAGGTGTGTTTGC |
| Reverse | CTTCATCAAGGGAATGATGG | |
| β-actin | Forward | ACAGGCATTGTGATGGACTC |
| Reverse | AGAAGGAAGGCTGGAAAAGA |
| Sample | DPPH (IC50, μg/mL) | ABTS (IC50, μg/mL) | Contents | |
|---|---|---|---|---|
| Total Phenol (mg GA/g Extract) | Total Flavonoid (mg CE/g Extract) | |||
| A. dioicus | 66.96 ± 3.17 b | 30.56 ± 0.20 b | 127.39 ± 1.04 | 104.17 ± 3.31 |
| Ascorbic acid | 4.36 ± 0.32 c | 5.91 ± 0.11 c | - | - |
| Phenolics (mg/kg) | Chlorogenic Acid | Caffeic Acid | Ferulic Acid | Veratric Acid | Cinnamic Acid | Quercetin |
| 7.04 ± 1.10 c | 20.14 ± 0.52 b | 1.74 ± 0.87 d | 29.31 ± 4.26 b | 4.70 ± 0.86 c | 418.41 ± 7.26 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.Y.; Park, J.-Y.; Seo, H.T.; Seong, H.-A.; Ji, Y.-J.; Lee, S.E.; Seo, K.H.; Kim, H.D. Samnamul (Shoots of Aruncus dioicus) Inhibit Adipogenesis by Downregulating Adipocyte-Specific Transcription Factors in 3T3-L1 Adipocytes. Processes 2020, 8, 1576. https://doi.org/10.3390/pr8121576
Lee JY, Park J-Y, Seo HT, Seong H-A, Ji Y-J, Lee SE, Seo KH, Kim HD. Samnamul (Shoots of Aruncus dioicus) Inhibit Adipogenesis by Downregulating Adipocyte-Specific Transcription Factors in 3T3-L1 Adipocytes. Processes. 2020; 8(12):1576. https://doi.org/10.3390/pr8121576
Chicago/Turabian StyleLee, Ji Yeon, Jeong-Yong Park, Hyun Taek Seo, Hyun-A Seong, Yun-Jeong Ji, Seung Eun Lee, Kyung Hye Seo, and Hyung Don Kim. 2020. "Samnamul (Shoots of Aruncus dioicus) Inhibit Adipogenesis by Downregulating Adipocyte-Specific Transcription Factors in 3T3-L1 Adipocytes" Processes 8, no. 12: 1576. https://doi.org/10.3390/pr8121576





