VectorDisk: A Microfluidic Platform Integrating Diagnostic Markers for Evidence-Based Mosquito Control
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mosquito Assays
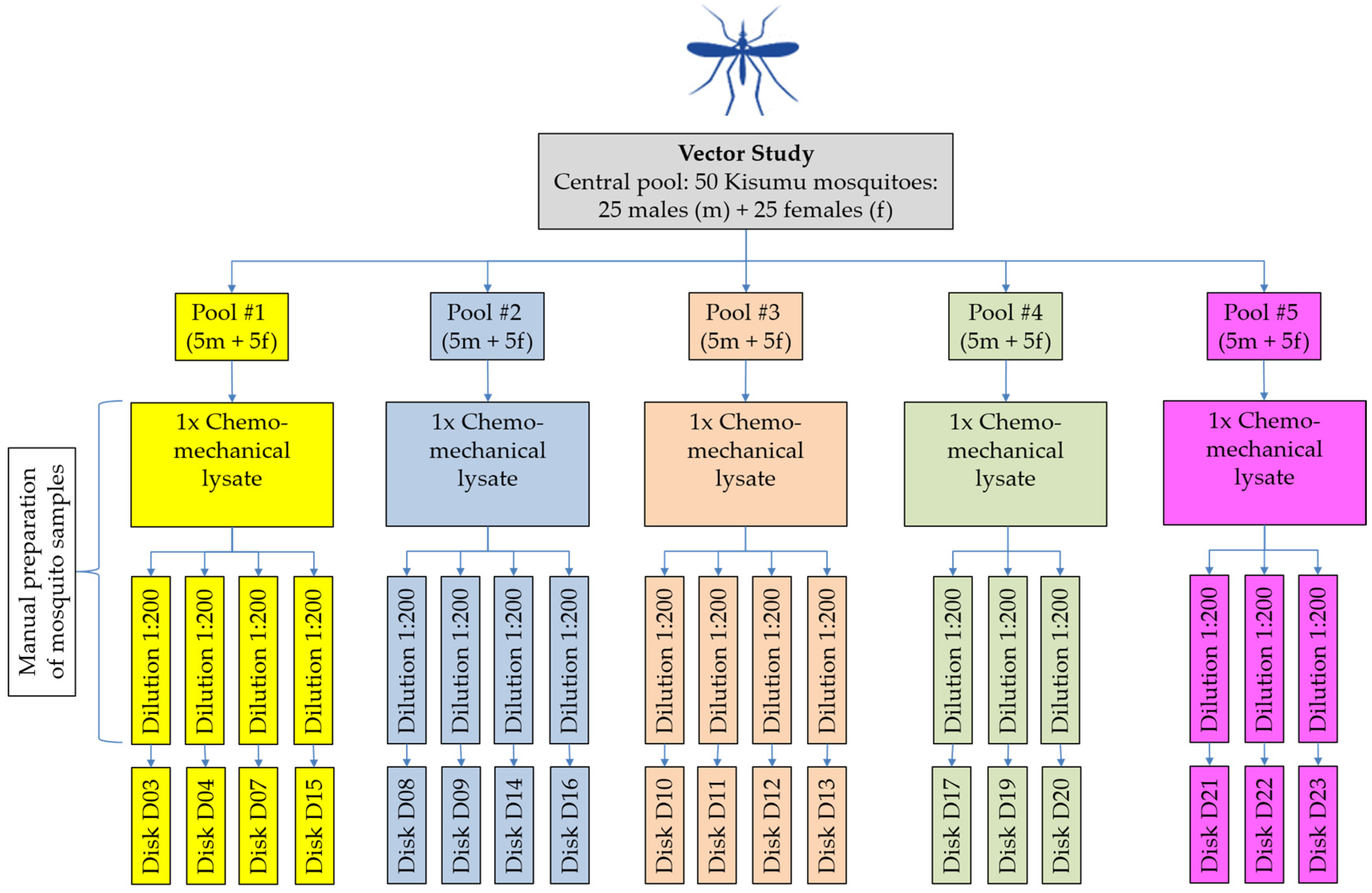
2.2. Vector Study: Manual Preparation of Mosquito Samples (Lysis and Homogenization)
2.3. Nucleic Acid Study
2.4. VectorDisk Fabrication
2.5. VectorDisk Workflow
3. Results and Discussion
3.1. The Vector Study Results
3.2. The Nucleic Acid Study Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Global Vector Control Response 2017–2030; World Health Organization (WHO): Geneva, Switzerland, 2017; Available online: https://www.who.int/vector-control/publications/global-control-response/en (accessed on 18 November 2020).
- Müller, R.; Reuss, F.; Kendrovski, V.; Montag, D. Vector-borne diseases. In Biodiversity and Health in the Face of Climate Change; Marselle, M., Stadler, J., Korn, H., Irvine, K., Bonn, A., Eds.; Springer: Cham, Switzerland, 2019; pp. 67–90. [Google Scholar] [CrossRef] [Green Version]
- Vorou, R. Zika virus, vectors, reservoirs, amplifying hosts, and their potential to spread worldwide: What we know and what we should investigate urgently. Int. J. Infect. Dis. 2016, 48, 85–90. [Google Scholar] [CrossRef] [Green Version]
- Heyman, P.; Cochez, C.; Hofhuis, A.; van der Giessen, J.; Sprong, H.; Porter, S.R.; Losson, B.; Saegerman, C.; Donoso-Mantke, O.; Niedrig, M.; et al. A clear and present danger: Tick-borne diseases in Europe. Expert Rev. Anti Infect. Ther. 2010, 8, 33–50. [Google Scholar] [CrossRef]
- Rochlin, I.; Ninivaggi, D.V.; Hutchinson, M.L.; Farajollahi, A. Climate change and range expansion of the Asian Tiger Mosquito (Aedes albopictus) in northeastern USA: Implications for public health practitioners. PLoS ONE 2013, 8, e60874. [Google Scholar] [CrossRef]
- Medlock, J.M.; Hansford, K.M.; Schaffner, F.; Versteirt, V.; Hendrickx, G.; Zeller, H.; Van Bortel, W. A review of the invasive mosquitoes in Europe: Ecology, public health risks, and control options. Vector Borne Zoonotic Dis. 2012, 12, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Rezza, G.; Nicoletti, L.; Angelini, R.; Romi, R.; Finarelli, A.C.; Panning, M.; Cordioli, P.; Fortuna, C.; Boros, S.; Magurano, F.; et al. Infection with chikungunya virus in Italy: An outbreak in a temperate region. Lancet 2007, 370, 1840–1846. [Google Scholar] [CrossRef]
- Venturi, G.; Di Luca, M.; Fortuna, C.; Remoli, M.E.; Riccardo, F.; Severini, F.; Toma, L.; Del Manso, M.; Benedetti, E.; Caporali, M.G.; et al. Detection of a chikungunya outbreak in Central Italy, August to September 2017. Eurosurveillance 2017, 22, 11–14. [Google Scholar] [CrossRef] [Green Version]
- Giron, S.; Franke, F.; Decoppet, A.; Cadiou, B.; Travaglini, T.; Thirion, L.; Durand, G.; Jeannin, C.; L’Ambert, G.; Grard, G.; et al. Vector-borne transmission of Zika virus in Europe, southern France, August 2019. Eurosurveillance 2019, 24, 2–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kioulos, I.; Kampouraki, A.; Morou, E.; Skavdis, G.; Vontas, J. Insecticide resistance status in the major West Nile virus vector Culex pipiens from Greece. Pest Manag. Sci. 2014, 70, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Pervanidou, D.; Vakali, A.; Georgakopoulou, T.; Panagiotopoulos, T.; Patsoula, E.; Koliopoulos, G.; Politis, C.; Stamoulis, K.; Gavana, E.; Pappa, S.; et al. West Nile virus in humans, Greece, 2018: The largest seasonal number of cases, 9 years after its emergence in the country. Eurosurveillance 2020, 25, 15–27. [Google Scholar] [CrossRef]
- Gossner, C.M.; Marrama, L.; Carson, M.; Allerberger, F.; Calistri, P.; Dilaveris, D.; Lecollinet, S.; Morgan, D.; Nowotny, N.; Paty, M.; et al. West Nile virus surveillance in Europe: Moving towards an integrated animal-human-vector approach. Eurosurveillance 2017, 22, 10–19. [Google Scholar] [CrossRef]
- Bhatt, S.; Weiss, D.J.; Cameron, E.; Bisanzio, D.; Mappin, B.; Dalrymple, U.; Battle, K.; Moyes, C.L.; Henry, A.; Eckhoff, P.A.; et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 2015, 526, 207–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranson, H.; Lissenden, N. Insecticide resistance in African Anopheles Mosquitoes: A worsening situation that needs urgent action to maintain malaria control. Trends Parasitol. 2015, 32, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Hemingway, J.; Field, L.; Vontas, J. An overview of insecticide resistance. Science 2002, 298, 96–97. [Google Scholar] [CrossRef] [PubMed]
- Vontas, J.; Mavridis, K. Vector population monitoring tools for insecticide resistance management: Myth or fact? Pest. Biochem. Physiol. 2019, 162, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Bass, C.; Williamson, M.S.; Field, L.M. Development of a multiplex real-time PCR assay for identification of members of the Anopheles gambiae species complex. Acta Trop. 2008, 107, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Bass, C.; Nikou, D.; Blagborough, A.M.; Vontas, J.; Sinden, R.E.; Williamson, M.S.; Field, L.M. PCR-based detection of Plasmodium in Anopheles mosquitoes: A comparison of a new high-throughput assay with existing methods. Malar. J. 2008, 7, 177. [Google Scholar] [CrossRef] [Green Version]
- Bass, C.; Nikou, D.; Vontas, J.; Donnelly, M.J.; Williamson, M.S.; Field, L.M. The vector population monitoring tool (VPMT): High-throughput DNA-based diagnostics for the monitoring of mosquito vector populations. Malar. Res. Treat. 2010, 2010, 190434. [Google Scholar] [CrossRef] [Green Version]
- Donnelly, M.J.; Isaacs, A.T.; Weetman, D. Identification, validation, and application of molecular diagnostics for insecticide resistance in malaria vectors. Trends Parasitol. 2016, 32, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Weetman, D.; Donnelly, M.J. Evolution of insecticide resistance diagnostics in malaria vectors. Trans. R. Soc. Trop. Med. Hyg. 2015, 109, 291–293. [Google Scholar] [CrossRef]
- Mitsakakis, K.; Hin, S.; Muller, P.; Wipf, N.; Thomsen, E.; Coleman, M.; Zengerle, R.; Vontas, J.; Mavridis, K. Converging human and malaria vector diagnostics with data management towards an integrated holistic one health approach. Int. J. Environ. Res. Public Health 2018, 15, 259. [Google Scholar] [CrossRef] [Green Version]
- WHO. Microscopy; World Health Organization: Geneva, Switzerland, 2017; Available online: www.who.int/malaria/areas/diagnosis/microscopy/en (accessed on 9 August 2020).
- Vontas, J.; Mitsakakis, K.; Zengerle, R.; Yewhalaw, D.; Sikaala, C.H.; Etang, J.; Fallani, M.; Carman, B.; Muller, P.; Chouaibou, M.; et al. Automated innovative diagnostic, data management and communication tool, for improving malaria vector control in endemic settings. Stud. Health Technol. Inform. 2016, 224, 54–60. [Google Scholar] [PubMed]
- Czilwik, G.; Messinger, T.; Strohmeier, O.; Wadle, S.; von Stetten, F.; Paust, N.; Roth, G.; Zengerle, R.; Saarinen, P.; Niittymaki, J.; et al. Rapid and fully automated bacterial pathogen detection on a centrifugal-microfluidic LabDisk using highly sensitive nested PCR with integrated sample preparation. Lab Chip 2015, 15, 3749–3759. [Google Scholar] [CrossRef] [PubMed]
- Hin, S.; Lopez-Jimena, B.; Bakheit, M.; Klein, V.; Stack, S.; Fall, C.; Sall, A.; Enan, K.; Frischmann, S.; Gillies, L.; et al. The FeverDisk: Multiplex detection of fever-causing pathogens for rapid diagnosis of tropical diseases. In Proceedings of the 21st International Conference on Miniaturized Systems for Chemistry and Life Sciences, μTAS 2017, Savannah, GA, USA, 22–26 October 2017; pp. 7–8. [Google Scholar]
- Rombach, M.; Hin, S.; Specht, M.; Johannsen, B.; Lüddecke, J.; Paust, N.; Zengerle, R.; Roux, L.; Sutcliffe, T.; Pecham, J.R.; et al. RespiDisk: A point-of-care platform for fully automated detection of respiratory tract infection pathogens in clinical samples. Analyst 2020, 145, 7040–7047. [Google Scholar] [CrossRef] [PubMed]
- Vernick, K.D. Infravec2: Expanding researcher access to insect vector tools and resources. Pathog. Glob. Health 2017, 111, 217–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kefi, M.; Mavridis, K.; Simoes, M.L.; Dimopoulos, G.; Siden-Kiamos, I.; Vontas, J. New rapid one-step PCR diagnostic assay for Plasmodium falciparum infective mosquitoes. Sci. Rep. 2018, 8, 1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bass, C.; Nikou, D.; Donnelly, M.J.; Williamson, M.S.; Ranson, H.; Ball, A.; Vontas, J.; Field, L.M. Detection of knockdown resistance (kdr) mutations in Anopheles gambiae: A comparison of two new high-throughput assays with existing methods. Malar. J. 2007, 6, 111. [Google Scholar] [CrossRef] [Green Version]
- Mavridis, K.; Wipf, N.; Müller, P.; Traore, M.M.; Muller, G.; Vontas, J. Detection and monitoring of insecticide resistance mutations in Anopheles gambiae: Individual vs pooled specimens. Genes (Basel) 2018, 9, 479. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.M.; Liyanapathirana, M.; Agossa, F.R.; Weetman, D.; Ranson, H.; Donnelly, M.J.; Wilding, C.S. Footprints of positive selection associated with a mutation (N1575Y) in the voltage-gated sodium channel of Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2012, 109, 6614–6619. [Google Scholar] [CrossRef] [Green Version]
- Bass, C.; Nikou, D.; Vontas, J.; Williamson, M.S.; Field, L.M. Development of high-throughput real-time PCR assays for the identification of insensitive acetylcholinesterase (ace-1R) in Anopheles gambiae. Pestic. Biochem. Physiol. 2010, 96, 80–85. [Google Scholar] [CrossRef]
- Mavridis, K.; Wipf, N.; Medves, S.; Erquiaga, I.; Müller, P.; Vontas, J. Rapid multiplex gene expression assays for monitoring metabolic resistance in the major malaria vector Anopheles gambiae. Parasites Vectors 2019, 12, 9. [Google Scholar] [CrossRef] [Green Version]
- Del Amo, J.; Sotelo, E.; Fernandez-Pinero, J.; Gallardo, C.; Llorente, F.; Aguero, M.; Jimenez-Clavero, M.A. A novel quantitative multiplex real-time RT-PCR for the simultaneous detection and differentiation of West Nile virus lineages 1 and 2, and of Usutu virus. J. Virol. Methods 2013, 2, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.W.; Yip, C.C.Y.; Tee, K.M.; Zhu, Z.; Tsang, J.O.L.; Chik, K.K.H.; Tsang, T.G.W.; Chan, C.C.S.; Poon, V.K.M.; Sridhar, S.; et al. Improved detection of Zika virus RNA in human and animal specimens by a novel, highly sensitive and specific real-time RT-PCR assay targeting the 5’-untranslated region of Zika virus. Trop. Med. Int. Health 2017, 22, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Hahn-Schickard Lab-on-a-Chip Foundry. Available online: https://www.hahn-schickard.de/en/production/lab-on-a-chip-foundry (accessed on 18 November 2020).
- Focke, M.; Stumpf, F.; Faltin, B.; Reith, P.; Bamarni, D.; Wadle, S.; Müller, C.; Reinecke, H.; Schrenzel, J.; Francois, P. Microstructuring of polymer films for sensitive genotyping by real-time PCR on a centrifugal microfluidic platform. Lab Chip 2010, 10, 2519–2526. [Google Scholar] [CrossRef] [PubMed]
- Focke, M.; Kosse, D.; Al-Bamerni, D.; Lutz, S.; Müller, C.; Reinecke, H.; Zengerle, R.; von Stetten, F. Microthermoforming of microfluidic substrates by soft lithography (µTSL): Optimization using design of experiments. J. Micromech. Microeng. 2011, 21, 115002. [Google Scholar] [CrossRef]
- Simmons, M.; Myers, T.; Guevara, C.; Jungkind, D.; Williams, M.; Houng, H.S. Development and validation of a quantitative, one-step, multiplex, real-time reverse transcriptase PCR assay for detection of dengue and Chikungunya viruses. J. Clin. Microbiol. 2016, 54, 1766–1773. [Google Scholar] [CrossRef] [Green Version]
- Kosse, D.; Buselmeier, D.; Müller, C.; Zengerle, Z.; von Stetten, F. Gas pressure assisted thermal bonding of film-based Lab-on-a-Chip cartridges. In Proceedings of the Mikrosystemtechnik-Kongress, Darmstadt, Germany, 10–12 October 2011; pp. 579–582. [Google Scholar]
- Keller, M.; Czilwik, G.; Schott, J.; Schwarz, I.; Dormanns, K.; von Stetten, F.; Zengerle, R.; Paust, N. Robust temperature change rate actuated valving and switching for highly integrated centrifugal microfluidics. Lab Chip 2017, 17, 864–875. [Google Scholar] [CrossRef]
- Zehnle, S.; Schwemmer, F.; Roth, G.; von Stetten, F.; Zengerle, R.; Paust, N. Centrifugo-dynamic inward pumping of liquids on a centrifugal microfluidic platform. Lab Chip 2012, 12, 5142–5145. [Google Scholar] [CrossRef] [Green Version]
- Hin, S.; Paust, N.; Keller, M.; Rombach, M.; Strohmeier, O.; Zengerle, R.; Mitsakakis, K. Temperature change rate actuated bubble mixing for homogeneous rehydration of dry pre-stored reagents in centrifugal microfluidics. Lab Chip 2018, 18, 362–370. [Google Scholar] [CrossRef]
- Burger, S.; Schulz, M.; von Stetten, F.; Zengerle, R.; Paust, N. Rigorous buoyancy driven bubble mixing for centrifugal microfluidics. Lab Chip 2016, 16, 261–268. [Google Scholar] [CrossRef]
- Mark, D.; Weber, P.; Lutz, S.; Focke, M.; Zengerle, R.; von Stetten, F. Aliquoting on the centrifugal microfluidic platform based on centrifugo-pneumatic valves. Microfluid. Nanofluid. 2011, 10, 1279–1288. [Google Scholar] [CrossRef]
- Strohmeier, O.; Keller, M.; Schwemmer, F.; Zehnle, S.; Mark, D.; von Stetten, F.; Zengerle, R.; Paust, N. Centrifugal microfluidic platforms: Advanced unit operations and applications. Chem. Soc. Rev. 2015, 44, 6187–6229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Johannsen, B.; Müller, L.; Baumgartner, D.; Karkossa, L.; Fruh, S.M.; Bostanci, N.; Karpisek, M.; Zengerle, R.; Paust, N.; Mitsakakis, K. Automated pre-analytic processing of whole saliva using magnet-beating for point-of-care protein biomarker analysis. Micromachines 2019, 10, 833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Jang, S.H.; Jia, G.Y.; Zoval, J.V.; Da Silva, N.A.; Madou, M.J. Cell lysis on a microfluidic CD (compact disc). Lab Chip 2004, 4, 516–522. [Google Scholar] [CrossRef]
- Kido, H.; Micic, M.; Smith, D.; Zoval, J.; Norton, J.; Madou, M. A novel, compact disk-like centrifugal microfluidics system for cell lysis and sample homogenization. Colloid Surf. B Biointerfaces 2007, 58, 44–51. [Google Scholar] [CrossRef]
- Siegrist, J.; Gorkin, R.; Bastien, M.; Stewart, G.; Peytavi, R.; Kido, H.; Bergeron, M.; Madou, M. Validation of a centrifugal microfluidic sample lysis and homogenization platform for nucleic acid extraction with clinical samples. Lab Chip 2010, 10, 363–371. [Google Scholar] [CrossRef]
- Hin, S.; Paust, N.; Rombach, M.; Lueddecke, J.; Specht, M.; Zengerle, R.; Mitsakakis, K. Minimizing ethanol carry-over in centrifugal microfluidic nucleic acid extraction by advanced bead handling and management of diffusive mass transfer. In Proceedings of the 20th International Conference on Solid-State Sensors, Actuators and Microsystems & Eurosensors XXXIII, Berlin, Germany, 23–27 June 2019; pp. 130–133. [Google Scholar] [CrossRef]
- Lopez-Jimena, B.; Bekaert, M.; Bakheit, M.; Frischmann, S.; Patel, P.; Simon-Loriere, E.; Lambrechts, L.; Duong, V.; Dussart, P.; Harold, G.; et al. Development and validation of four one-step real-time RT-LAMP assays for specific detection of each dengue virus serotype. PLoS Negl. Trop. Dis. 2018, 12, e0006381. [Google Scholar] [CrossRef]
- Lopez-Jimena, B.; Wehner, S.; Harold, G.; Bakheit, M.; Frischmann, S.; Bekaert, M.; Faye, O.; Sall, A.A.; Weidmann, M. Development of a single-tube one-step RT-LAMP assay to detect the Chikungunya virus genome. PLoS Negl. Trop. Dis. 2018, 12, e0006448. [Google Scholar] [CrossRef]






| Reaction Chamber | Assay Panel | Nucleic Acid Type | Assay Target (FAM) | Assay Target (HEX) | Assay Target (ATTO 647N) |
|---|---|---|---|---|---|
| 2 | Species ID | DNA | Aq 1 | Ag 1 | Aa 1 |
| 3 | Molecular Forms | DNA | S 2 | M 2 | - |
| 4 | Kdr | DNA | Rw 3 | S-wt 4 | Re 3 |
| 5 | Kdr+ | DNA | R | S-wt | - |
| 6 | iAChe | DNA | R | S-wt | - |
| 7 | Plasmodium species | DNA | P. falciparum | P. ovm5 | - |
| 8 | Infective stage | RNA | - | PLP1 | - |
| 9 | Detox (A) | RNA | RPS7 | CYP6P3 | CYP6M2 |
| 10 | Detox (B) | RNA | RPS7 | CYP9K1 | CYP6P4 |
| 11 | Detox (C) | RNA | RPS7 | CYP6Z1 | GSTE2 |
| 12 | Detox (D) | RNA | RPS7 | CYP6P1 | CYP4G16 |
| Reaction Chamber | Assay Panel | Nucleic Acid Type | Assay Target (FAM) | Assay Target (HEX) | Assay Target (ATTO 647N) |
|---|---|---|---|---|---|
| 2 | West Nile virus | RNA | WNV-Lineage 1 | WNV-Lineage 2 | - |
| 3 | Zika virus | RNA | - | - | ZIKV |
| 4 | Dengue virus | RNA | DENV T1-4 | - | - |
| 5 | Plasmodium species ID | DNA | P. falciparum | P. ovm1 | - |
| 6 | Plasmodium infective stage | RNA | - | - | Pf infective stage |
| 7 | West Nile virus | RNA | WNV-Lineage 1 | WNV-Lineage 2 | - |
| 8 | Zika virus | RNA | - | - | ZIKV |
| 9 | Dengue virus | RNA | DENV T1-4 | - | - |
| 10 | Plasmodium species ID | DNA | P. falciparum | P. ovm1 | - |
| 11 | Plasmodium infective stage | RNA | - | - | Pf infective stage |
| No. of Lysate Pool | Pool #1 | Pool #2 | Pool #3 | Pool #4 | Pool #5 | Overall | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Green detection channel | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | |
| 2 | Species ID (Aq) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 3 | Molecular Forms (S) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 4 | Kdr (Rw) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 5 | Kdr+ (R) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 6 | iAChe (R) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 7 | P. falciparum | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 8 | - | ||||||||||||||||||||||||
| 9 | Detox (A) RPS7 | 3/4 | 1/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 17/18 | 1/18 | ||||||||||||
| 10 | Detox (B) RPS7 | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 2/3 | 1/3 | 3/3 | 0/3 | 17/18 | 1/18 | ||||||||||||
| 11 | Detox (C) RPS7 | 4/4 | 0/4 | 3/4 | 1/4 | 4/4 | 0/4 | 2/3 | 1/3 | 3/3 | 0/3 | 16/18 | 2/18 | ||||||||||||
| 12 | Detox (D) RPS7 | 4/4 | 0/4 | 4/4 | 0/4 | 3/4 | 1/4 | 3/3 | 0/3 | 3/3 | 0/3 | 17/18 | 1/18 | ||||||||||||
| Yellow detection channel | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | |
| 2 | Species ID (Ag) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 3 | Molecular Forms (M) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 4 | kdr (S-wt) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 5 | kdr+ (S-wt) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 6 | iAChE (S-wt) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 7 | Plasmodium ovm | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 8 | Infective stage (PLP1) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 9 | Detox (A) CYP6P3 | 3/4 | 1/4 | 4/4 | 0/4 | 3/4 | 1/4 | 3/3 | 0/3 | 3/3 | 0/3 | 16/18 | 2/18 | ||||||||||||
| 10 | Detox (B) CYP9K1 | 1/4 | 3/4 | 2/4 | 2/4 | 2/4 | 2/4 | 2/3 | 1/3 | 1/3 | 2/3 | 8/18 | 10/18 | ||||||||||||
| 11 | Detox (C) CYP6Z1 | 4/4 | 0/4 | 2/4 | 2/4 | 3/4 | 1/4 | 1/3 | 2/3 | 3/3 | 0/3 | 13/18 | 5/18 | ||||||||||||
| 12 | Detox (D) CYP6P1 | 3/4 | 1/4 | 1/4 | 3/4 | 3/4 | 1/4 | 3/3 | 0/3 | 1/3 | 2/3 | 11/18 | 7/18 | ||||||||||||
| Red detection channel | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | |
| 2 | Species ID (Aa) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 3 | - | ||||||||||||||||||||||||
| 4 | Kdr (Re) | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 | 3/3 | 0/3 | 3/3 | 0/3 | 18/18 | 0/18 | ||||||||||||
| 5 | - | ||||||||||||||||||||||||
| 6 | - | ||||||||||||||||||||||||
| 7 | - | ||||||||||||||||||||||||
| 8 | - | ||||||||||||||||||||||||
| 9 | Detox (A) CYP6M2 | 1/4 | 3/4 | 4/4 | 0/4 | 2/4 | 2/4 | 0/3 | 3/3 | 3/3 | 0/3 | 10/18 | 8/18 | ||||||||||||
| 10 | Detox (B) CYP6P4 | 0/4 | 4/4 | 0/4 | 4/4 | 1/4 | 3/4 | 0/3 | 3/3 | 1/3 | 2/3 | 2/18 | 16/18 | ||||||||||||
| 11 | Detox (C) GSTE2 | 4/4 | 0/4 | 2/4 | 2/4 | 3/4 | 1/4 | 0/3 | 3/3 | 2/3 | 1/3 | 11/18 | 7/18 | ||||||||||||
| 12 | Detox (D) CYP4G16 | 3/4 | 1/4 | 2/4 | 2/4 | 3/4 | 1/4 | 3/3 | 0/3 | 0/3 | 3/3 | 11/18 | 7/18 | ||||||||||||
| Sample | WNV-Lineage 1 | ZIKV | PF-INF | Overall | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Green detection channel | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | |
| 2 | WNV-Lineage 1 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 6/6 | 0/6 | ||||||
| 3 | - | ||||||||||||||||
| 4 | DENV T1-4 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 9/9 | 0/9 | ||||||||
| 5 | Plasmodium falciparum | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 6/6 | 0/6 | ||||||
| 6 | - | ||||||||||||||||
| 7 | WNV-Lineage 1 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 6/6 | 0/6 | ||||||
| 8 | - | ||||||||||||||||
| 9 | DENV T1-4 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 9/9 | 0/9 | ||||||||
| 10 | Plasmodium falciparum | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 6/6 | 0/6 | ||||||
| 11 | - | ||||||||||||||||
| Yellow detection channel | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | |
| 2 | WNV-Lineage 2 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 9/9 | 0/9 | ||||||||
| 3 | - | ||||||||||||||||
| 4 | - | ||||||||||||||||
| 5 | Plasmodium ovm1 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 9/9 | 0/9 | ||||||||
| 6 | - | ||||||||||||||||
| 7 | WNV-Lineage 2 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 9/9 | 0/9 | ||||||||
| 8 | - | ||||||||||||||||
| 9 | - | ||||||||||||||||
| 10 | Plasmodium ovm1 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 9/9 | 0/9 | ||||||||
| 11 | - | ||||||||||||||||
| Red detection channel | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | TP | FN | TN | FP | |
| 2 | - | ||||||||||||||||
| 3 | ZIKV | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 6/6 | 0/6 | ||||||
| 4 | - | ||||||||||||||||
| 5 | - | ||||||||||||||||
| 6 | Pf infective stage | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 6/6 | 0/6 | ||||||
| 7 | - | ||||||||||||||||
| 8 | ZIKV | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 6/6 | 0/6 | ||||||
| 9 | - | ||||||||||||||||
| 10 | - | ||||||||||||||||
| 11 | Pf infective stage | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 3/3 | 0/3 | 6/6 | 0/6 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hin, S.; Baumgartner, D.; Specht, M.; Lüddecke, J.; Mahmodi Arjmand, E.; Johannsen, B.; Schiedel, L.; Rombach, M.; Paust, N.; von Stetten, F.; et al. VectorDisk: A Microfluidic Platform Integrating Diagnostic Markers for Evidence-Based Mosquito Control. Processes 2020, 8, 1677. https://doi.org/10.3390/pr8121677
Hin S, Baumgartner D, Specht M, Lüddecke J, Mahmodi Arjmand E, Johannsen B, Schiedel L, Rombach M, Paust N, von Stetten F, et al. VectorDisk: A Microfluidic Platform Integrating Diagnostic Markers for Evidence-Based Mosquito Control. Processes. 2020; 8(12):1677. https://doi.org/10.3390/pr8121677
Chicago/Turabian StyleHin, Sebastian, Desirée Baumgartner, Mara Specht, Jan Lüddecke, Ehsan Mahmodi Arjmand, Benita Johannsen, Larissa Schiedel, Markus Rombach, Nils Paust, Felix von Stetten, and et al. 2020. "VectorDisk: A Microfluidic Platform Integrating Diagnostic Markers for Evidence-Based Mosquito Control" Processes 8, no. 12: 1677. https://doi.org/10.3390/pr8121677
APA StyleHin, S., Baumgartner, D., Specht, M., Lüddecke, J., Mahmodi Arjmand, E., Johannsen, B., Schiedel, L., Rombach, M., Paust, N., von Stetten, F., Zengerle, R., Wipf, N., Müller, P., Mavridis, K., Vontas, J., & Mitsakakis, K. (2020). VectorDisk: A Microfluidic Platform Integrating Diagnostic Markers for Evidence-Based Mosquito Control. Processes, 8(12), 1677. https://doi.org/10.3390/pr8121677





