Abstract
In mammalian cell culture, especially in pharmaceutical manufacturing, pH is a critical process parameter that has to be controlled as accurately as possible. Not only does pH directly affect cell culture performance, ensuring a comparable pH is also crucial for scaling and transfer of processes. A sample-based offline pH measurement is commonly used to ensure correct bioreactor pH probe signals after sterilization and as a detection measure for drifts of probe signals. However, the sample-based pH offline measurement does not necessarily deliver required accuracy. Offsets between bioreactor pH and sample pH heavily depend on equipment, local procedures and the offline measurement method that is used. This article adequately describes a novel, non-invasive method to determine pH and pCO2 in sterile bioreactors without the need to sample and measure offline. This method utilizes the chemical correlation between carbon dioxide in the gas phase, dissolved carbon dioxide, bicarbonate and dependent proton concentrations that directly affect the pH in carbonate buffered systems. The proposed carbon dioxide-based pH reference method thereby is able to accurately determine the true pH in the bioreactor without the need to sample. The proposed method is independent of scale and bioreactor configuration and does not depend on local procedures that may differ between sites, scales or operators. Applicability of the method for both stainless steel and single use bioreactors is shown. Furthermore, the very same principles are applicable for non-invasive, online pCO2 monitoring.
1. Introduction
Mammalian cell culture processes in a pharmaceutical environment usually take place in tightly controlled bioreactors to ensure comparable process performance, productivity and product quality. One parameter of particular interest is the pH value, and we agree that there are not necessarily consensus guidelines for best practice in managing pH in cell cultures. Furthermore, reporting standards relating to pH are typically inadequate [1]. This applies especially for publications of both academics and industry impeding efforts to reproduce findings.
The current standard for monitoring and control of bioreactor pH-probe signals in biotechnological processes relies on sample-based offline methods (e.g., pH-meters and connected glass electrodes or blood gas analyzers). Typically, bioreactor pH-probe signals are frequently compared to pH values that are derived by sample-based offline measurement. Additionally, bioreactor probe signals are not considered accurate after sterilization. If the difference exceeds defined criteria, some kind of readjustment strategy is applied, considering the offline value the source of truth. After adjusting the bioreactor pH-probe signal onto the sample-based offline value, bioreactor pH is a representation of the local offline measurement.
Sampling procedures, equipment, sample hold times, shifts in sample temperature and carbon dioxide degassing all invariably add offsets, leading to a different pH in the sample compared to the true pH in the bioreactor. Avoiding those offsets is thereby impossible. Keeping offsets constant between scales or sites is hardly achievable. Especially at higher cell densities, pH must not be considered stable after sampling. Furthermore, offsets must not be considered consistent during a fermentation even if sample hold times are kept comparable because media buffering, pCO2 contents, and cell densities all change. Furthermore, sample temperatures change after sampling, depending on sample equipment and procedures. Although blood gas analyzers typically maintain 37 °C, this is not necessarily the case if pH meters are used. This also is relevant for calibration procedures that may take place at temperatures differing from sample temperatures. Temperature compensation does thereby not account for temperature dependency of pH in different solutions.
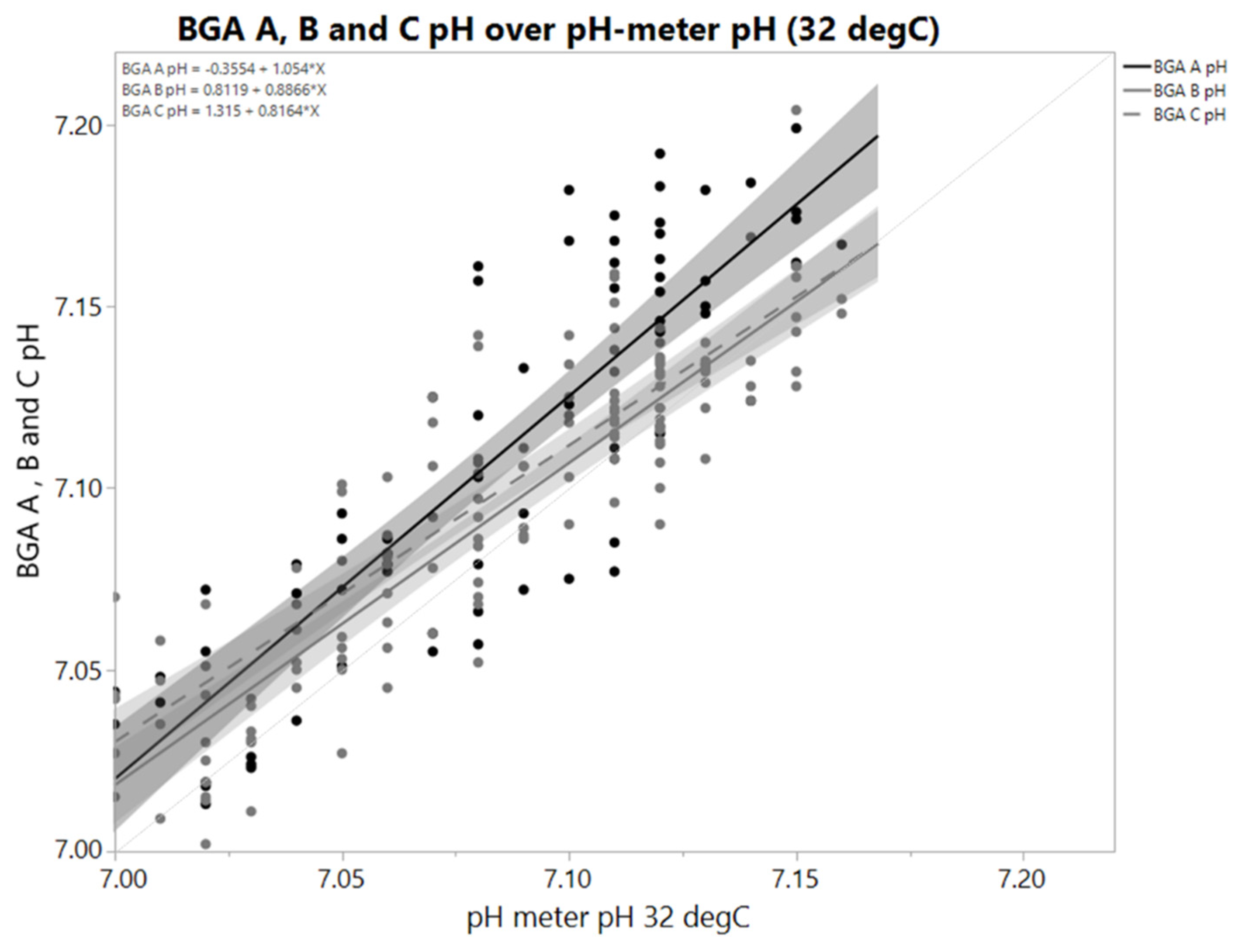
Blood gas analyzers as well as pH-meters are commonly used for offline measurement. pH values derived from blood gas analyzers potentially differ from pH values derived by pH-meters [2]. This also holds true if samples from controlled systems as bioreactors are measured (Figure 1). A sample temperature of 32 ± 2 °C is thereby maintained in all scales to match the at-scale sample temperature if pH-meters are used. Assessing the difference between a certain reference (e.g., bioreactor pH or another pH measurement) reveals a high spread. An offset might be different for the next sample-based offline reading because calibration data, operators, sample properties and so on might have changed between sample time points. The overall information content of a sample-based offline measurement is questionable. Correlations of pH derived by offline measurements to parameters directly affected by pH are often weak or not significant, although process characterization usually proves pH to be a critical process parameter affecting those.
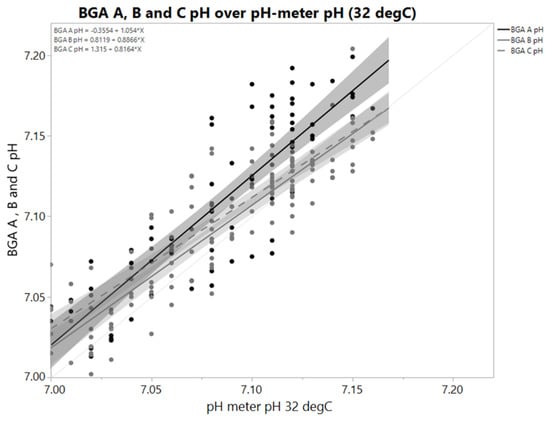
Figure 1.
pH values derived by a pH-meter and three blood gas analyzers A, B and C in identical samples; where B and C are two comparable devices. X-axis shows pH-meter value. Sample temperature for pH meter was controlled at 32 °C ± 2 °C. Samples came from eight parallel bioreactor runs (N = 72). Temperature of measurement chamber of all blood gas analyzers was 37 °C. Grey areas are 95% confidence intervals of the linear regression. pH decreases with increasing temperature, so difference between BGA pH and pH-meter cannot be explained by a temperature effect. Slopes and offsets both differ. That means difference depends on either actual pH or sample properties that correlate with pH. Spread of differences is high. A single pH value obviously may not contain required information to justify a readjustment step.
If local procedures introduce different offsets, a pH of 7.00 at one site may simply mean something other than 7.00 at another site. Unknown pH variability thereby manifests in parameters that directly or indirectly depend on pH such as growth, lactate generation, product concentration and certain quality attributes. The amount of base needed to maintain pH obviously directly depends on pH as well, increasing e.g., osmolality.
To enable a more accurate pH control and to ensure more comparable pH values after scaling or transfer of cell culture processes, a method to determine pH without sampling is highly beneficial.
This also holds true when reproducing published findings is not possible due to insufficient description of sampling procedures, equipment, offline measurement methods, sample hold times and readjustment strategies based on that offline measurement. It is insufficient to report a pH set point alone or together with the offline method.
Mammalian cell culture media are usually carbonate buffered. Cells in uncontrolled systems like shake flasks, T-flasks or micro titer plates therefore are typically cultivated in incubators that allow a controlled carbon dioxide concentration to maintain pH values in desired ranges. Depending on the bicarbonate and carbon dioxide concentrations in the gas phase, the corresponding pH can be calculated or predicted using nomograms or respective tables.
The ratios of the respective species, carbon dioxide, carbonic acid and bicarbonate in aqueous solutions as well as resulting pCO2 and pH, have been published in satisfactory detail elsewhere and are usually part of academic training [3,4]. It is very important to note that not only pH and carbon dioxide in the gas phase must correlate. Since carbon dioxide in the gas phase and dissolved carbon dioxide are directly related via the same chemical principles, we also must be able to estimate dissolved carbon dioxide (pCO2) with off-gas analysis. However, to match sample-based offline pCO2 values and the actual pCO2 in the bioreactor, sample hold times had to be included as well due to changes in carbon dioxide concentration in closed sample containers after sampling before measurement. This means that like pH, pCO2 derived by sample-based offline measurement may not be representative of the true value in bioreactors, partly via the very same mechanisms like carbon dioxide degassing during the sampling process or carbon dioxide accumulation via respiration. The potential to estimate pCO2 levels and trends via off-gas analysis is also discussed in this paper very briefly.
Interrelations between pH, pCO2, bicarbonate and dissolved carbon dioxide are not dependent on scale or, e.g., bioreactor configuration. pH will be identical in identical medium at a given carbon dioxide concentration in the gas phase no matter what volume a given shake flask has, or if that is compared to the pH in a T-flask in the same incubator. This is true in any system including bioreactors, where carbon dioxide is used as an acidic component for pH control, counteracting any carbon dioxide removal by aeration of this bioreactor.
We can assume that in equilibrium, flux of exhaust carbon dioxide equals carbon dioxide influx. Under this assumption, overall CO2 mass in the bioreactor must be constant. Given constant pressure and temperature, dissolved carbon dioxide and bicarbonate concentration and pH must be constant as well.
This means pH in a given cell culture medium can be calculated if carbon dioxide concentration in a respective bioreactor gas phase is known, and if both carbon dioxide concentration and pH are kept stable, e.g., via pH control or defined gas influx containing carbon dioxide. This is also true not only for stainless steel bioreactors, but also for single use bioreactors. Technical implementation of off-gas analysis itself might differ depending on the bioreactor used, but underlying principles stay the same [5]. The correlation of pH and corresponding carbon dioxide concentration in the gas phase is thereby universal and applicable, independent of location and the vessel used.
Nonetheless, in GMP manufacturing the independence of this method from scale and bioreactor configuration has to be proven and validated against the standard established method, which is sample-based offline measurement. As described, sample-based offline pH measurement comes with inherent variability. To validate the off-gas-based pH reference method, we still have to refer to the very same sample-based offline measurement that we want to replace by a more accurate method.
An unintended shift in pH after changing to the proposed method in legacy products therefore needs to be avoided. The proposed method is able to determine the true pH in the bioreactor without sampling and offsets introduced by the sample-based offline method. On the other hand, we are currently using sample-based offline measurement to monitor and readjust our bioreactor pH-probes. Offsets currently introduced by sample-based offline measurement must therefore be accurately determined. Either pH set points have to be adapted to match the true pH, or the offsets have to be incorporated in the method itself.
In 2004, the U.S. Department of Health and Human Services Food and Drug Administration published a framework for Innovative Pharmaceutical Development, Manufacturing, and Quality Assurance, emphasizing the need for applying scientific and engineering principles to ensure acceptable and reproducible product quality in manufacturing processes. Thereby, pH must be considered a relevant parameter affecting product quality and overall process performance in mammalian cell culture. Any significant advance in the ability to get this parameter better and more accurately controlled might have a positive impact on overall process control and in the end patient safety.
Furthermore, reproducing findings that have been achieved somewhere else in the scientific community would finally be possible as well in the field of mammalian cell culture where different cell lines, proprietary media and insufficient description of materials and methods slow down potential overall progress.
2. Materials and Methods
2.1. Determination of Media Specific Correlations of the pH and Corresponding Exhaust Carbon Dioxide Concentration
To determine the exhaust carbon dioxide concentration, an off-gas measurement was established (DASGIP® GA4 (Eppendorf AG, Hamburg, Germany). Carbon dioxide concentration was determined with double beam infrared absorption. Pressure compensation was inactive; environmental pressure was set to 941 mbar (Roche Site in Penzberg is 621 m in height). The CO2 sensor was two point calibrated before use with process air and a defined gas mixture (Linde AG) containing 24% CO2 and 2% O2.
Two-Liter Bioreactors (Sartorius B-DCU Quad) with a total volume of 3 L and typical working volumes of around 2 L were connected to the off-gas sensors with gas tight tubing. For experimental determination of media specific correlations of pH and a corresponding exhaust carbon dioxide concentration up to 2.5 L, medium was used to minimize headspace volume. Pressure control valve, sterile filter and exhaust cooling at around ten °C were integrated in the exhaust line. Pressure was controlled to match at-scale total pressures for respective products. Bioreactors and pH probes (Mettler Toledo, InPro3253/225/PT100) were sterilized wet in 1.2 g/L KH2PO4(aq). Media fill was performed under sterile conditions.
Bioreactor pH was determined with a built in online-pH probe that was initially two point calibrated in buffers (4.00 and 7.00 at 25 °C, Mettler Toledo). Carbon dioxide gas was used as an acidic pH correction agent via pH control to maintain pH at upper dead bands. The pH controller was set up as a proportional controller to achieve a constant carbon dioxide influx. No basic correction agent was used, and so pH thereby naturally increases by carbon dioxide removal via constant aeration with process air.
After sterilization and media fill a hold step to stabilize pH, pressure and temperature were established. To enable accurate pH readings, a port in the bioreactor lid was opened, making the system unsterile in the process. Two independent pH-probes (Mettler Toledo InLab Semi-Micro) connected to respective pH-meters (Knick Portavo 907) were inserted simultaneously through the open port into the bioreactor to measure pH in the liquid phase without sampling. Agitation, aeration and temperature control all stayed active. Pressure control was set to inactive. pH-meters were independently three point calibrated (Mettler Toledo buffers 9.21, 7.00, 4.00 at 25 °C ) with active automated temperature compensation (ATC). The average of the pH-meter readings was used to standardize (one point adjustment) the online bioreactor pH probe signal. Maximum acceptable difference of the pH meter signals was thereby 0.02, and the maximum difference from the buffer pH was 0.01. The same procedure was performed to check bioreactor pH after the experiment again to detect any unintended drifts of the bioreactor probe signal.
After standardization of the bioreactor probe signal, the bioreactor lid was closed again and pressure control was set as active. After establishing an equilibrium (stable pH and carbon dioxide concentration in the exhaust gas) the first data point online pH and corresponding exhaust carbon dioxide concentration was documented. Correlations were determined in four technical replicates, which means four independent bioreactors filled with identical medium. Four pH set points were determined in each bioreactor. After establishing an equilibrium at every set point, pH was also determined via a sample-based offline measurement (Sarstedt Monovette, Knick Portavo 970 with connected Mettler Toledo InLab Semi-Micro Pro pH probe). Sample temperature thereby was kept at 32 ± 2 °C using appropriate sample tempering.
The cell culture medium shown in this paper is a bicarbonate buffered, complex proprietary medium.
2.2. Transfer and Use of Correlations in GMP Manufacturing
Correlations were fitted with quadratic regression (JMP 12). For transfer purposes, the equation was transferred to a new plant or site. All bioreactors in GMP manufacturing or clinical plants were equipped with two redundant, independent pH probes. Readjustment of bioreactor probes after media fill before inoculation was based on the result of this equation after actual carbon dioxide concentration was fed into the equation. The calculated pH was then used to initially standardize respective bioreactor probe signals. To detect potential bioreactor pH probe signal drifts, the readjustment strategy was adjusted to rely on delta probe alarms as a detection measure instead of frequent sample-based offline readings. Readjustments based on sample-based offline measurements only took place if a justified suspicion for a double probe drift that is not detectable by a delta probe alarm did exist. In case of a single probe drift detectable by a delta probe alarm an offline reading was utilized to determine which one of the probes measured incorrectly. A readjustment onto the offline value was thereby avoided at all times.
To establish an accurate reading, the carbonate buffered medium in the bioreactor has to be in equilibrium with its gas phase, which means either the pH control adds as much carbon dioxide as is removed via aeration or a defined influx containing carbon dioxide is used leading to stable readings of pH and corresponding carbon dioxide in both cases. Depending on scale and the ratio between headspace and working volume, the time until equilibrium may take more time then feasible. Aeration rates were adapted in those cases to accelerate headspace back mixing.
Additional parameters potentially adding errors like overpressure, headspace aeration, exhaust humidity and condensate, overall gas flow rates and so on were kept identical or adapted accordingly.
2.3. Validation of Scale Independency and Offset Determination to Established Sample-Based Offline Measurement
To prove independence of scale in an established commercial process, a correlation pH and corresponding exhaust carbon dioxide concentration were determined on a small scale as described above. Scale independency was proven by using the very same correlation in another scale (20 L) to adjust the bioreactor probe signal onto a calculated pH based on this correlation. After adjustment, the bioreactor was again made unsterile to directly measure with accurately calibrated pH-meters in the liquid phase of that bioreactor to prove the true pH to be comparable given the defined acceptance criteria.
To prove that the proposed method is as good or better as the established sample-based offline pH in scale up to 5k, pH values were calculated for each scale for the respective product using the quadratic fit for the media specific correlation. Equivalence of the offsets between sample-based offline values and the calculated pH was then tested using a two-sided student’s t-test (TOST). As practically significant differences (PSDs), the double standard deviation of historic differences of sample-based offline measurement and bioreactor pH was calculated. Equivalence is thereby considered proof that sample-based offline measurement does not generate different offsets on different scales due to different procedures and equipment.
2.4. Influence of pH Correction Agents Added during Media Preparation
Media preparation was considered with respect to weighing and dosing tolerances. pH correction agents especially directly affect the correlation pH and corresponding carbon dioxide concentration in consecutive bioreactors where those media are used. The lower the lot-to-lot variability is, the higher the accuracy of the carbon dioxide-based pH reference method will be.
Respective cell culture medium was filled in bioreactors (Sartorius B-DCU Quad). pH control was set as active, and no basic solution attached. The aeration rate was set constant to 50 ccm. After reaching equilibrium, where pH is maintained at the upper dead band by a constant carbon dioxide influx applied by pH control, the first data point pH and corresponding exhaust carbon dioxide concentration were documented. After adding a defined amount of pH correction agent, the next data point pH and corresponding exhaust carbon dioxide concentration were documented. Two technical replicates were used for two media lots to determine the difference in exhaust carbon dioxide readings at identical bioreactor pH caused by base addition.
Generally, for comparable media, comparable amounts of pH correction agent should be required. If media pH is adjusted to desired pH levels based on pH readings generated in the very same media by varying amounts of pH correction agents, conditions in bioreactors filled with those media cannot be achieved because the pH controller response, such as adding base and acid, will be altered. Furthermore, the very important process control (pH in finished medium) is annulled if media pH is adjusted because pH will always be in range after adjustment.
All projects so far that utilized the proposed method internally implemented fixed amounts of pH correction agents.
2.5. Exhaust Gas Analysis in Single Use Bioreactors (SUBs)
To determine carbon dioxide concentration in the exhaust gas of SUBs, DASGIP® GA4 (Eppendorf AG, Hamburg, Germany) devices were used. Before use, sensors were two point calibrated with process air and a defined gas mixture (Linde AG) containing 2% oxygen, 10% carbon dioxide (up to 250 L non GMP runs) and 24% carbon dioxide (2K GMP runs), respectively, to ensure a stable gas composition measurement at given ambient conditions. A membrane pump was connected after respective exhaust filters to ensure a stable and pulsation free exhaust gas flux from 250 L and 2k single use bioreactors w/o pressure control. Exhaust filter-heating prevented blocking. Condensate traps in the exhaust line removed any condensate generated by cooling of the saturated exhaust gas stream after filters.
All Ambr250™ multi fermenter systems (Sartorius Stedim Biotech AG, Göttingen, Germany) have a built-in off-gas analyzer module to measure O2 and CO2 in the exhaust flow of all 12 bioreactor vessels simultaneously with sufficient accuracy. CO2 sensors in Ambr250 were five-point calibrated with different defined gas mixtures (Linde AG) to reach calibration points at 0%, 3%, 5%, 8% and 16% CO2.
In all runs performed in SUBs, recombinant CHO suspension cell lines expressing different types of antibodies were cultivated in one culture medium (proprietary chemically defined Roche medium).
For standardization of the bioreactor pH-probe signal, respective media was subsequently equilibrated with a defined gas mixture (93% process air and 7% CO2) after media fill until saturation. After equilibration phase, culture pH was calculated from the exhaust carbon dioxide concentration in all scales based on the calibration function according to the described method above. Bioreactor probe signal was then adjusted onto the calculated pH.
Sample-based offline measurement was performed with varying methods depending on vessel size. Ambr250™ built-in analysis module was two point calibrated (Sartorius Stedim buffers 4.00, 7.00 at 25 °C ). A liquid handling system (200 µL) did perform sampling and offline pH measurement.
On the 250 L SUB scale, samples were drawn using a sterile syringe and pH was offline measured by a calibrated (Nova Biomedical calibration cartridge) blood gas analyzer (BioProfile® pHOx® Analyzer, Nova Biomedical GmbH, Mörfelden-Walldorf, Germany).
Samples in 2k SUBs in a GMP environment were also drawn using a sterile syringe and transferred into a preheated falcon tube. Offline pH was measured by a two point adjusted glass electrode (Knick Portavo 970 with connected Mettler Toledo Semi Micro pH probe, Mettler Toledo buffers 7.00, 4.00 at 25 °C ).
2.6. Exhaust Gas Analysis for Monitoring of Dissolved Carbon Dioxide
A recombinant CHO clone (Chinese Hamster Ovary cells, suspension culture) constitutively expressing an antibody was cultivated in proprietary chemically-defined cell culture media. All cell cultivations used for calibration of the pCO2 model took place in a 2 L stirred tank bioreactor (BIOSTAT® B-DCU Quad, Sartorius, Göttingen) for approximately 14 days cultivation time. Temperature, DO, pressure and pH were all controlled and kept constant for all runs shown.
Daily sample-based offline pCO2 measurement samples were drawn using a closed and tempered (32 °C ± 2 °C, Sarstedt Monovette) container to minimize unintended carbon dioxide. pCO2 was determined by a blood gas analyzer (cobas b 221, Roche Custom Biotech Mannheim, Germany).
Exhaust carbon dioxide concentration again was determined as described above (DASGIP® GA4 (Eppendorf AG, Hamburg, Germany).
The data set for developing a suitable model included a total of 32 cell cultivations. Calculations and modelling was carried out using JMP 12 (SAS, Cary, NC, USA) and Microsoft Excel 2016 (Redmond, WA, USA). The data set was split into a calibration set, containing 24 of the runs, and an external validation set comprising the remaining runs (1/4 of the complete data set). The model contained the actual exhaust carbon dioxide concentration as a quadratic effect, and actual pH, as equilibrium constants of the carbonate buffer systems depend on pH [4] and sample hold times to account for carbon dioxide changes in the sample until measurement. Those changes can easily be detected by calculating the ratio of pCO2 and exhaust carbon dioxide concentration. Exhaust carbon dioxide concentration does not change by sampling, whereas pCO2 in the sample does change. Looking at the ratio allows one to detect pCO2 changes in a sample by simply plotting it against a sample hold time, whereas measuring multiple times in one sample would potentially affect the outcome. Sample hold times were estimated.
A multilinear regression generic model was set up. This model thereby is only applicable for comparable sampling procedures, sampling equipment, offline method and the process conditions used. Sample hold times thereby have a relatively small effect and could be dismissed without altering the general outcome.
Equation (1): generic model to estimate sample-based offline pCO2 [%] by exhaust carbon dioxide [%], bioreactor pH and sample hold time [minutes]:
3. Results
3.1. Media Specific Correlations of the pH and Corresponding Carbon Dioxide Concentration
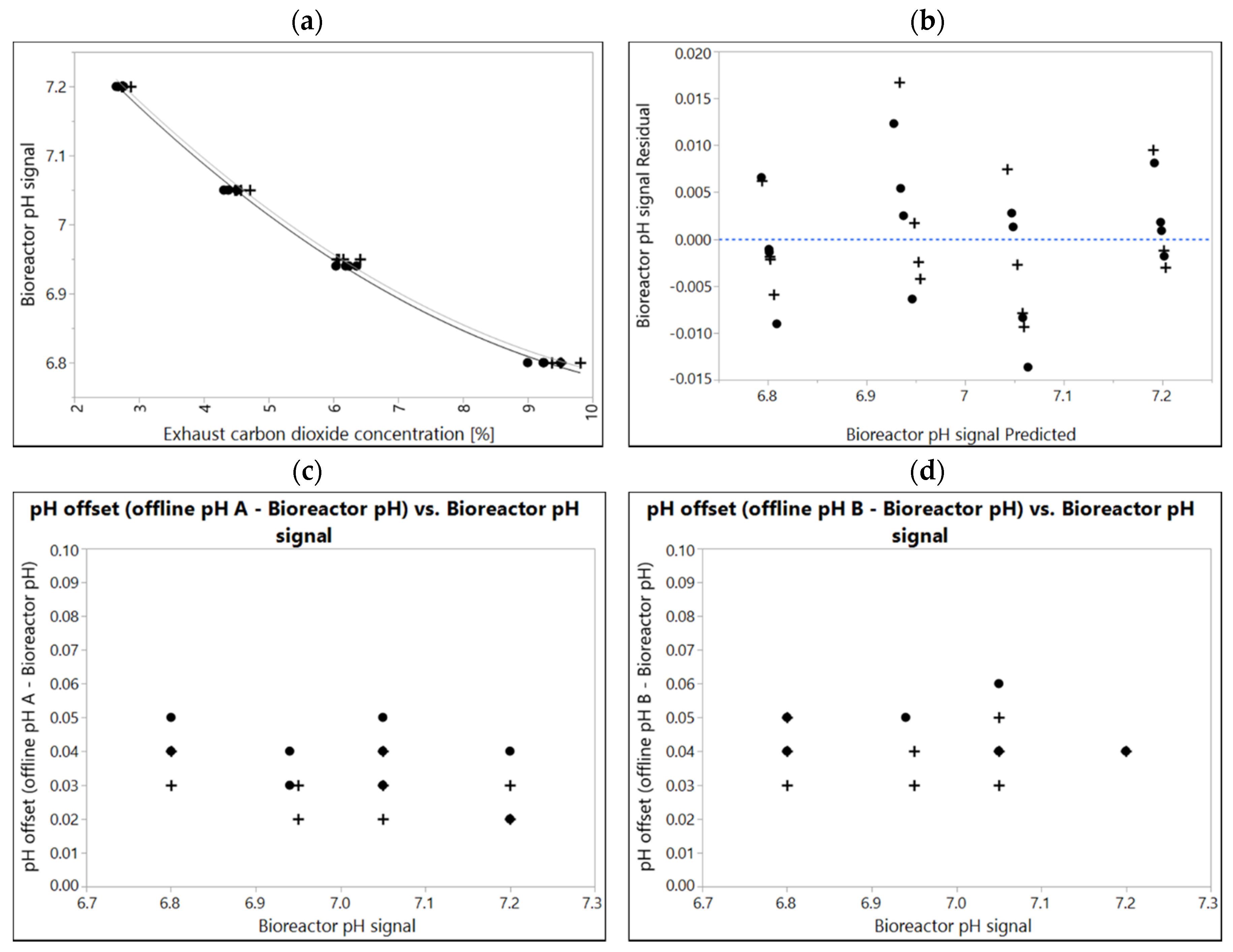
Media specific correlations of the two independent production media lots manufactured (10k scale) are shown in Figure 2. Although the difference between individually fitted correlations is significant (p = 0.00222), the difference in the resulting pH calculated from each correlation was <0.01. For this respective product, data points from two media lots were fitted together, resulting in residuals <0.02 for each data point from the fit model. Lot-to-lot variability was considered negligible in this case.
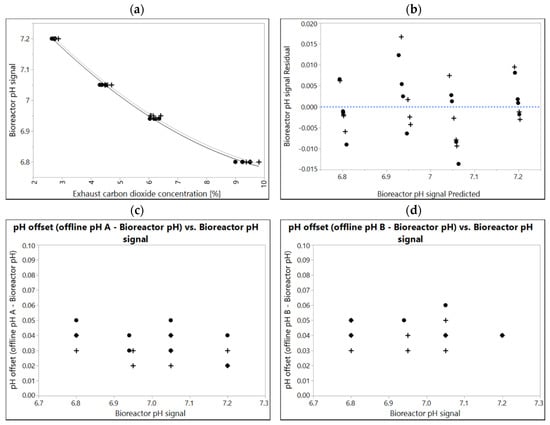
Figure 2.
(a) A correlation of bioreactor pH and corresponding exhaust carbon dioxide concentration in two different media lots manufactured independently (10k scale). Correlations of lot A (black dots) and lot B (plusses) were experimentally determined in 2 L, N = 4 at different points in time (two weeks apart). The line and dotted line are quadratic fits. Overpressure 300 mbar, Temperature 37 °C. If data of two independent experiments given in (a) are fitted together, residuals of combined fit (b) still range between ± 0.02 pH. (c,d) difference sample-based offline value from bioreactor pH ranging between 0.02 and 0.06 determined with two independent pH-meters.
Equation (2): quadratic equation to calculate pH with the exhaust carbon dioxide concentration (CO2, [%]) in equilibrium Media Lot A [%] rounded:
Equation (3): quadratic equation to calculate pH with the exhaust carbon dioxide concentration (CO2 [%]) in equilibrium Media Lot B [%] rounded:
For each data point a sample-based offline pH value has also been determined (Figure 2a,b). Calibration procedure for the pH meters is stricter here than criteria for most products allow. Furthermore, conditions for sampling and sample-based offline measurement are usually more defined in small-scale labs vs. large-scale manufacturing. Therefore, offsets shown here may be considered best case, and highlight the maximum accuracy that can be achieved using sample-based offline measurement in this case. All offsets are positive. This is typical in cell free media and can be explained by temperature (measurements were performed at 32 ± 2 °C compared to 37 °C in the bioreactor) as well as carbon dioxide degassing effects during sampling, sample hold time and measurement. Although pH meters were calibrated in identical buffers and were measuring identical samples, individual data points may already differ by up to 0.03 pH.
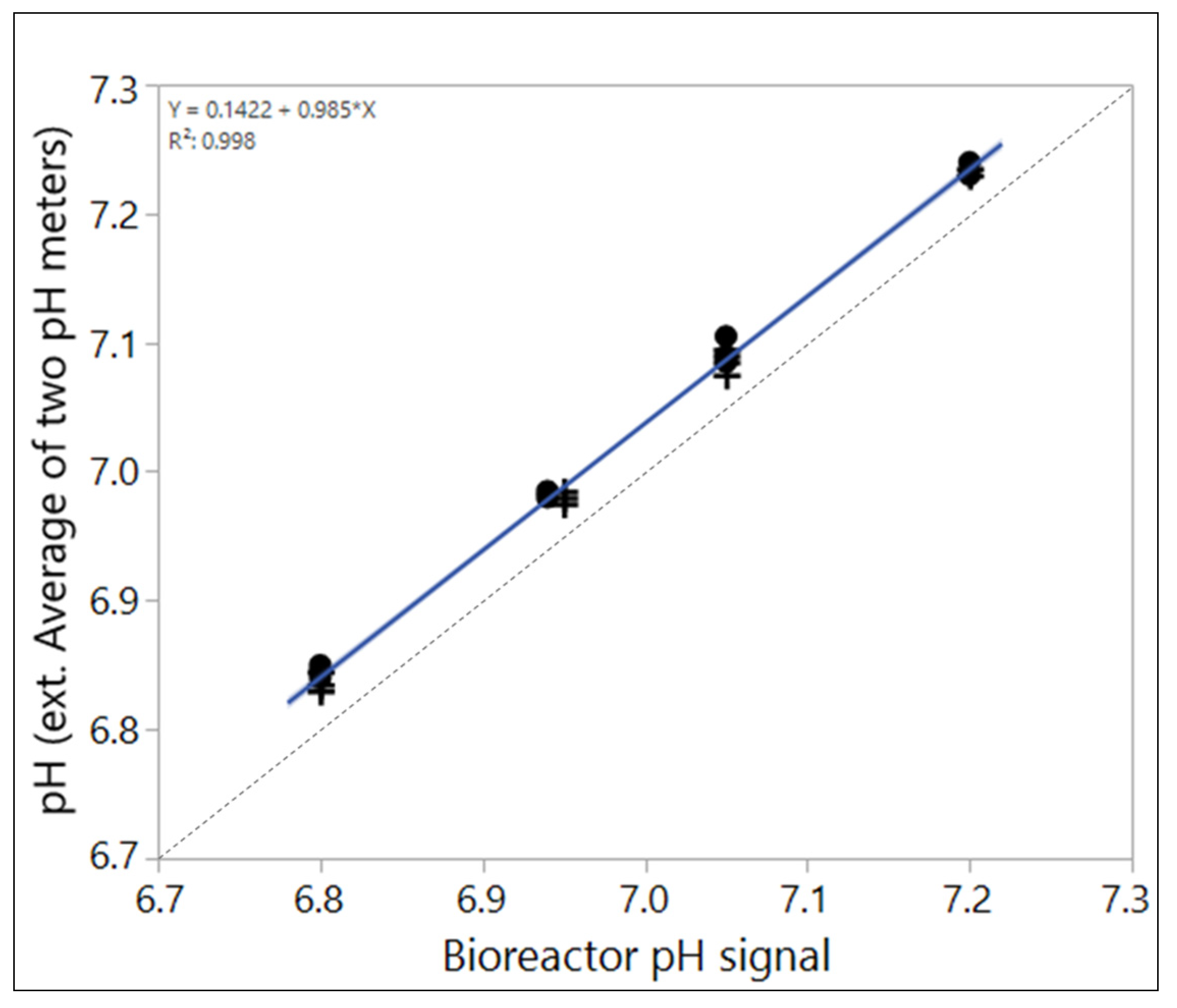
Although one single offline value may not represent the true bioreactor pH, it is possible to estimate an average offset to the true pH (Figure 3). This may be especially helpful to determine local offsets that may vary between sites for transfer purposes. The higher the variability of the respective local offsets between sample-based offline measurements and the true pH of the cell environment, the higher the risk of making a GMP decision (like readjusting bioreactor pH-probe signal) that is based on a flawed reading. Averages therefore have to be considered with care, depending on the implemented pH readjustment strategy.
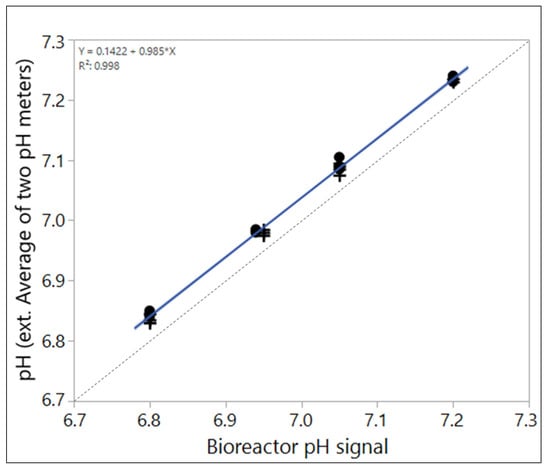
Figure 3.
Average of sample-based offline values of two pH-meters compared to the bioreactor pH signal that reflects the true pH in the case shown in Figure 2. Clearly, an offset is detectable that averages around 0.04 pH in this case.
3.2. Effect of pH Correction Agent on Media Specific Correlations of the pH and Corresponding Carbon Dioxide Concentration
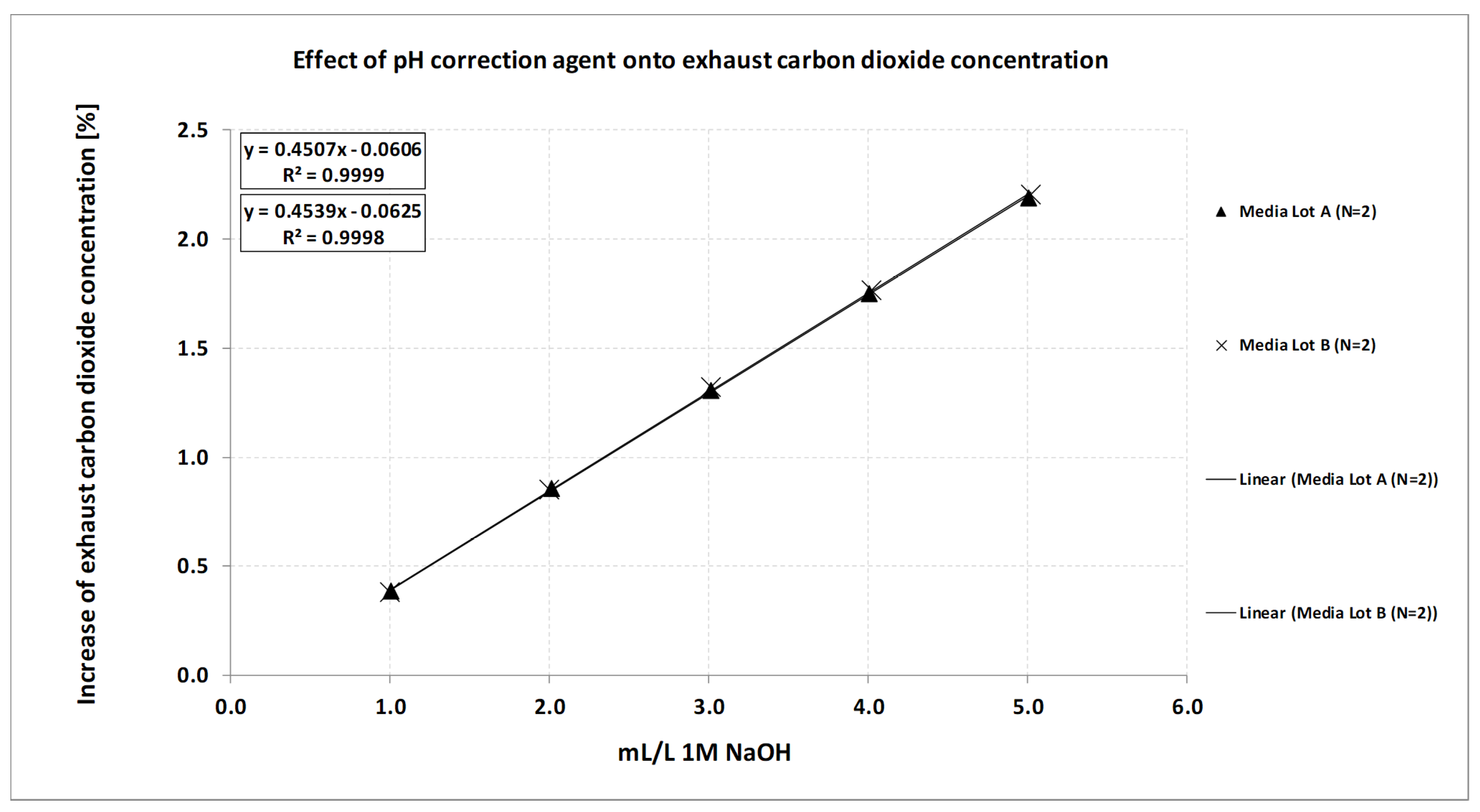
Media preparation procedures directly affect correlations like the ones shown in Figure 2a. The most relevant ingredients are solutions at extreme pH or the pH correction agent itself. Dosing and weighing tolerances may add some additional variability as well. The effect of the pH correction agent on the exhaust carbon dioxide concentration at a given pH in consecutive bioreactors where pH is controlled via carbon dioxide gas as the acidic component is shown in Figure 4. In this case, an increase of around 0.5% carbon dioxide in the exhaust would result in a pH difference of around 0.03 pH in this medium given the correlation shown in Figure 2a. If the effect of correction agent is well known, acceptance criteria can be derived to assess if the method is feasible even if media preparation itself introduces some errors. When a certain correlation is used to calculate pH from the exhaust carbon dioxide concentration, varying amounts of pH correction agent that are added during media preparation to counteract, e.g., carbon dioxide removal will add error to the method.
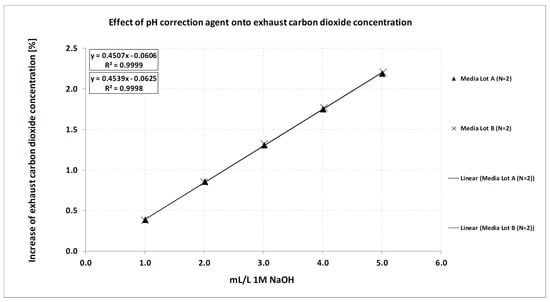
Figure 4.
Effect of pH correction agent 1 M NaOH on the exhaust carbon dioxide concentration in 2 L bioreactor systems with active pH control and carbon dioxide gas as the acidic component. In the range tested, the correlation is linear and almost identical between media lots.
3.3. Determination of Offsets between Sample-Based Offline Measurement and Carbon Dioxide Derived pH for Validation
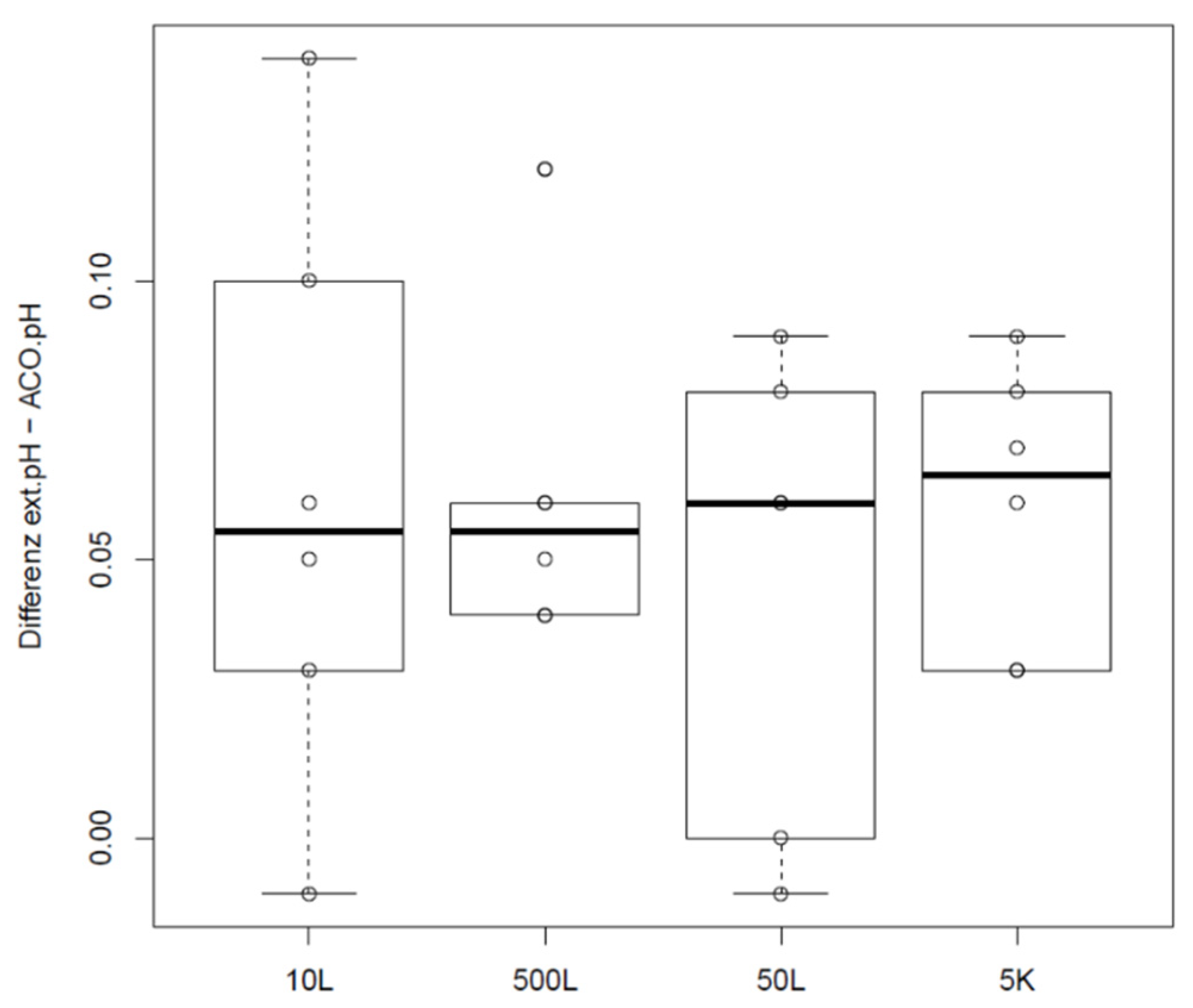
Offsets and distributions of the difference between sample-based offline measurement and the pH derived by a correlation pH and corresponding exhaust carbon dioxide concentration are shown in Figure 5. Data were determined in a GMP manufacturing line for a legacy product with a production scale of 5000 L as part of the validation efforts for this product. Scale independency for this product was shown in 20 L as described in Materials and Methods. Furthermore, equivalence of the offsets between sample-based offline values and bioreactor pH in scales 10 L, 50 L, 500 L and 5000 L were proven with two one sided t-tests (TOST) using double the standard deviation of historic differences between online and offline pH values as practically significant differences.
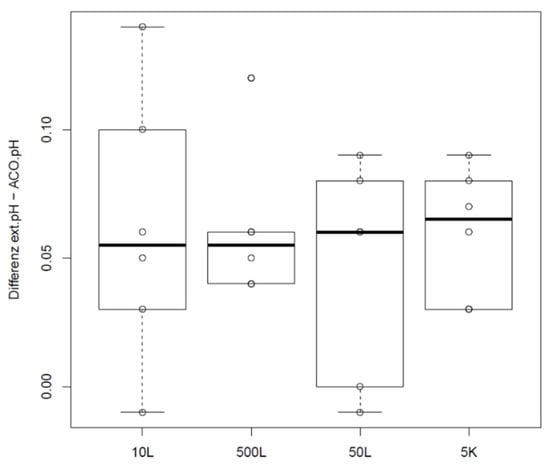
Figure 5.
Difference of sample-based offline values and pH calculated from the exhaust carbon dioxide concentration depending on scale.
Offsets introduced by sample-based offline measurements obviously do not differ significantly between scales in this case. It must be noted that even if respective mean difference is within the defined PSDs, offset of a single data point may vary tremendously in reality. Compared to the offset and variability shown for the small scale data in Figure 1 and Figure 2, it becomes clear that sampling and calibration procedures on the manufacturing floor may introduce offsets and variability that affects process performance. If the decision to readjust bioreactor probes is based on one single sample-based offline reading, true pH may already vary by about 0.1 pH, looking at this dataset alone.
3.4. Feasibility of the Carbon Dioxide-Derived pH in Single Use Bioreactors
The time for the equilibration phase depended on vessel size, headspace to working volume ratio and aeration rates and did partially take up to 9 h on a 2K scale.
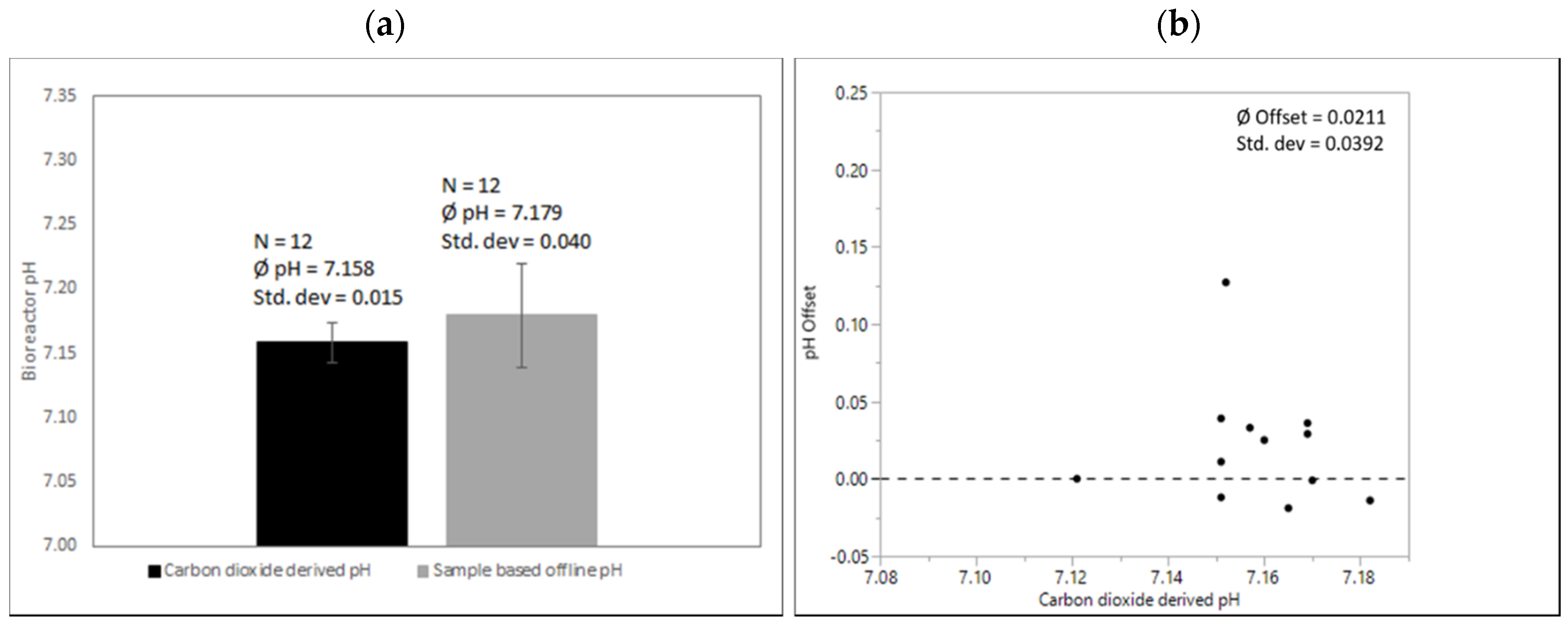
Scalability and particularly the robustness of this method are shown in Figure 6. pH in all runs in all scales was calculated from a quadratic fit generated on a small scale as described above. Figure 6 compares pH values derived from exhaust carbon dioxide concentration vs. sample-based offline measurement in 250 L SUBs.
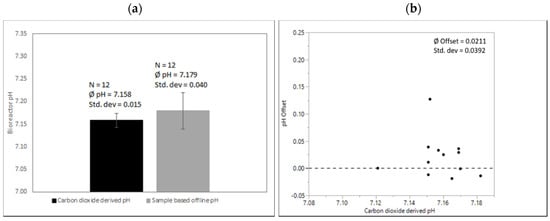
Figure 6.
(a) Comparison of carbon dioxide-derived and sample-based pH before inoculation after media fill. pH-probe signals were adjusted onto correlation exhaust carbon dioxide concentration and corresponding pH in all 250 L SUBs. Processes were executed from various operators over a period of several months with the same culture media recipe and the same conditions at the calibration time point as described above. (b) pH offset of sample-based offline values compared to quadratic fit for correlation exhaust carbon dioxide concentration and corresponding pH.
Again, an almost negligible mean difference between the sample-based and carbon dioxide-derived pH can be observed (0.021). However, the standard deviation of the sample-based offline measurement compared to the proposed method again reflects a high variability. This highlights that mean offsets might differ depending on local procedures, but all sample-based offline methods potentially add a high pH variability to cell culture operations. An offset of >0.1 for a single data point is not unusual.
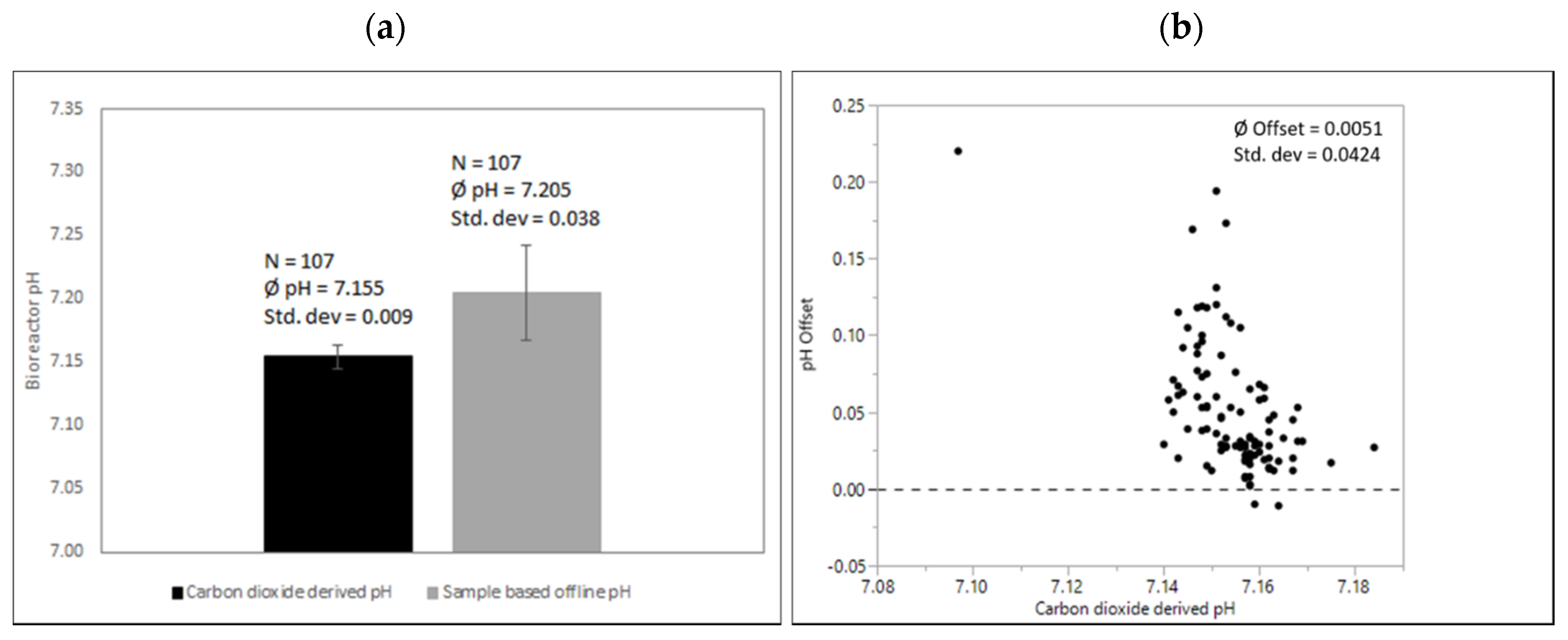
Sample-based offline measurements via automated liquid handling in Ambr250™ systems compared to pH derived by exhaust carbon dioxide concentrations are shown in Figure 7.
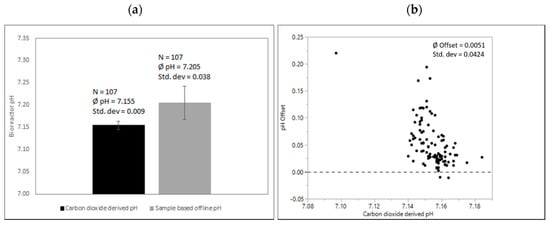
Figure 7.
(a) Comparison of carbon dioxide-derived and sample-based pH before inoculation after media fill. Processes were executed from various operators over a period of several months with the same culture media recipe and the same conditions at the calibration time point as described. (b) pH offsets of sample-based offline values compared to quadratic fit for correlation exhaust carbon dioxide concentration and corresponding pH. Due to its small volume and automated handling, exposure to carbon dioxide free environmental conditions is considered high.
As mentioned, sample exposition to a CO2-free environment has to be considered high, impacting pH via carbon dioxide degassing. Offsets were mainly positive with a mean difference of around 0.05. Again, variability is high adding variability to cell culture operations if single data points are used to adjust respective bioreactor pH-probe signals.
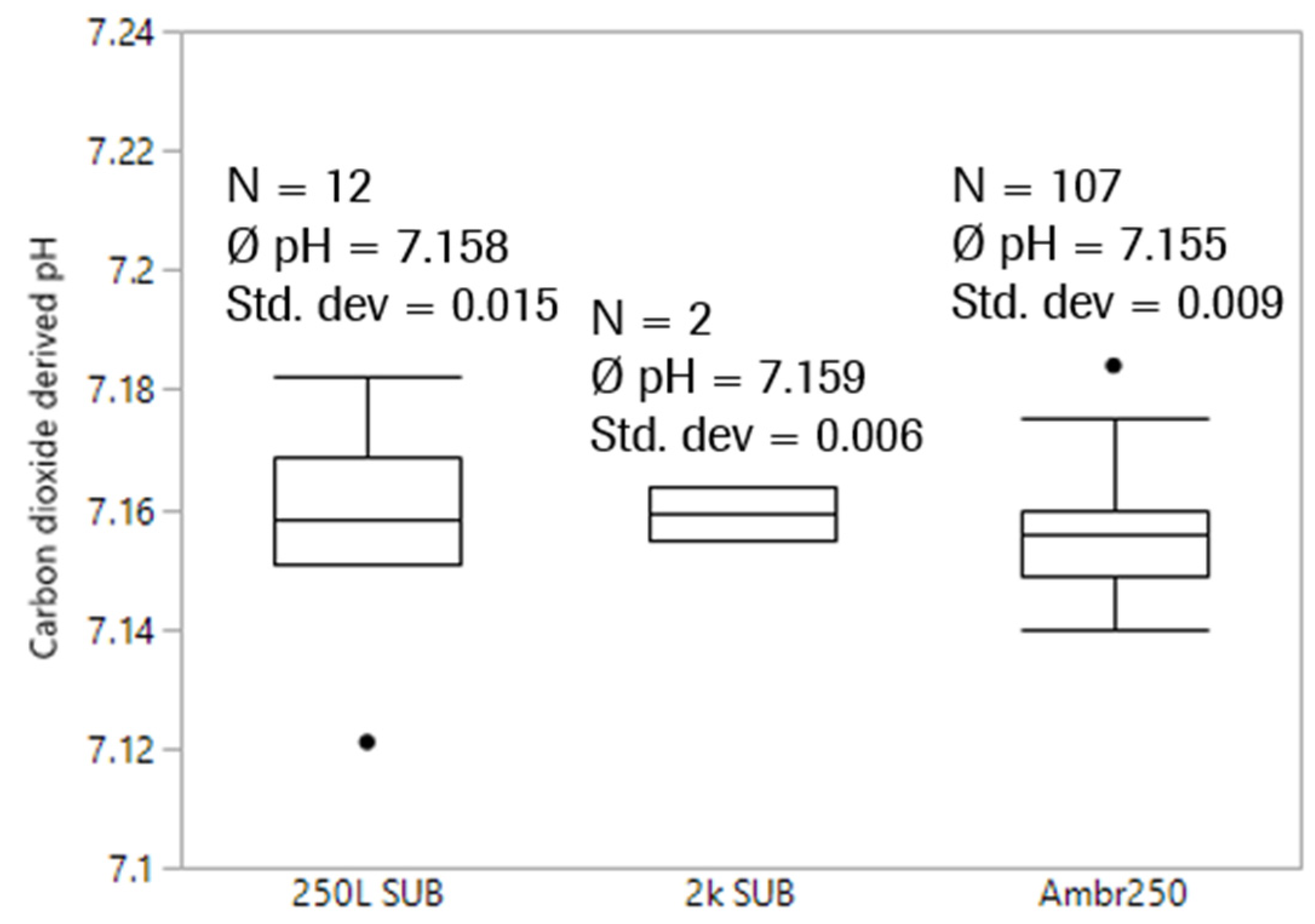
After transferring the quadratic fit formula into clinical GMP manufacturing, carbon dioxide concentrations of two 2k SUBs with more or less comparable media fill and equilibration procedures were determined and the pH was calculated (Figure 8).
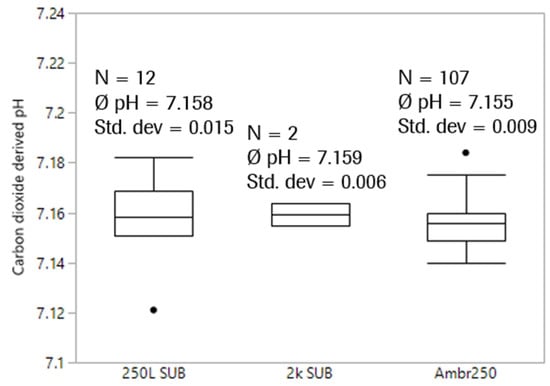
Figure 8.
Comparison of carbon dioxide-derived pH after media fill before inoculation between scales and plants. pH offline values for 2k SUBs had an offset of 0.009 and 0.046, respectively.
pH values derived from the exhaust carbon dioxide concentration are obviously very comparable and show low variability. Although many potential sources of error exist including media lot-to-lot differences, environmental pressure changes, slight differences in pressure and temperature control to name a few, knocking out offsets by avoiding sampling and sample-based offline measurement does affect comparability and precision of pH measurement using the proposed method.
3.5. Online pCO2 Monitoring Potential
Trends of the raw exhaust CO2 concentration measured by the off-gas analyzer as well as pCO2 modelled as described above are shown in Figure 9. Sample-based offline pCO2 measurements using a blood gas analyzer are shown as well. All systems had active pressure and temperature control with identical set points.
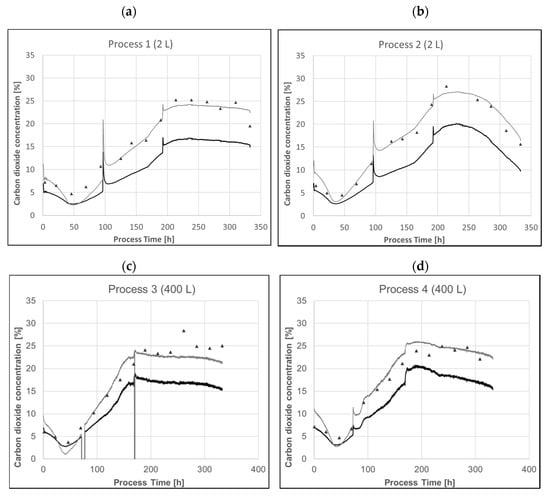
Figure 9.
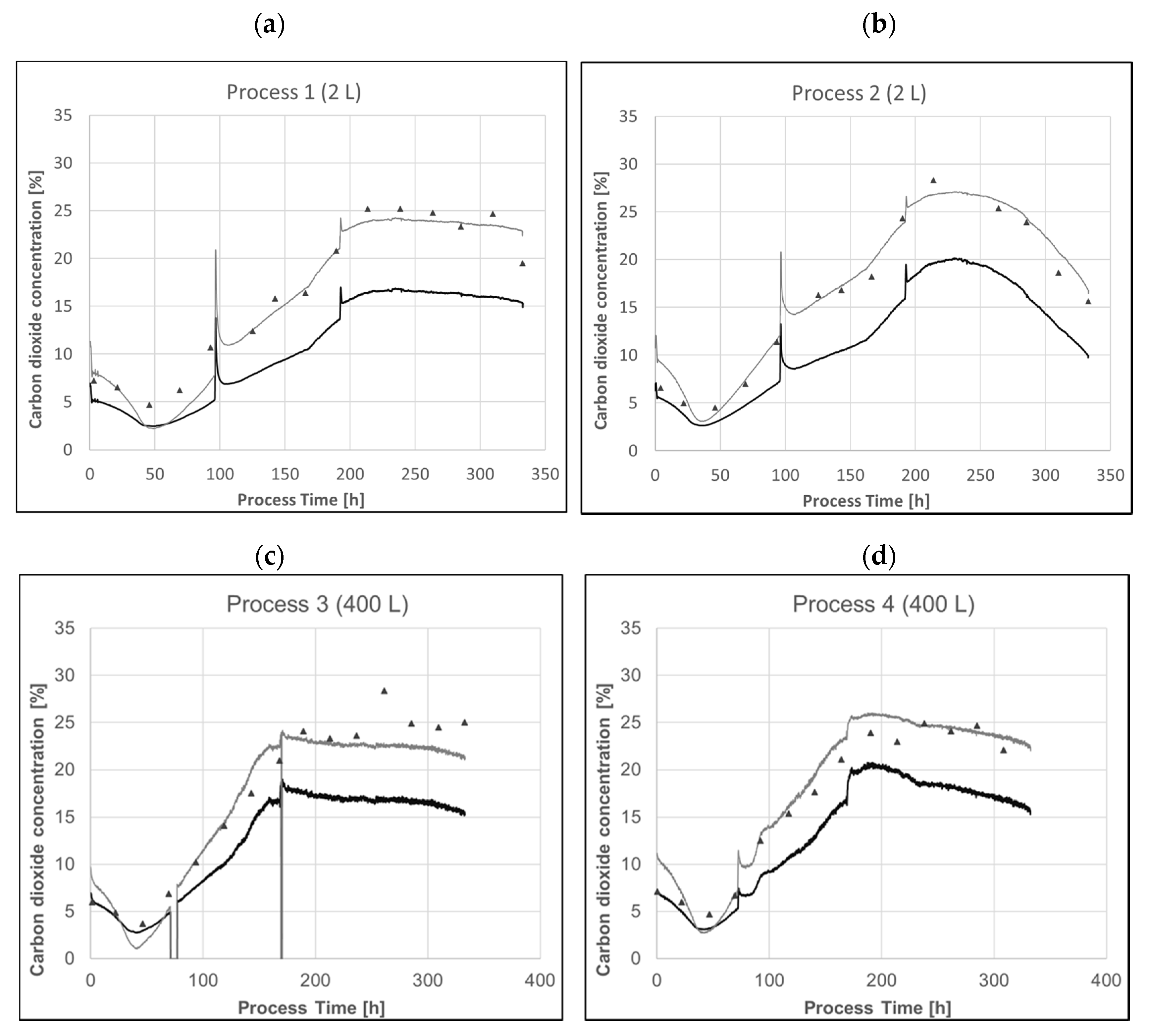
(a,b) Profiles of raw exhaust CO2 concentration (black line), predicted pCO2 concentration using Equation (1) with a sample hold time of 3 min (a) or 4.5 min (b) (grey line) and the sample-based pCO2 measurement (black triangles) of a representative process in a 2 L system. (c,d) Profiles of exhaust CO2 concentration (black line), of predicted pCO2 concentration using Equation (1) with a sample hold time of 1 min (grey line) of a representative process in a 400 L system. The model was derived from 2 L data and directly applied to 400 L. All bioreactors were pressure controlled at 400 mbar overpressure.
Exhaust-gas analysis can predict offline pCO2 relatively accurately with very little effort. Applying the model derived from 2 L data to 400 L runs show the potential for scalability and transferability of those models. Even using the raw carbon dioxide signal represents the trend very well. The main driver for the raw data not directly representing pCO2 is the applied overpressure. The system pressure directly affects mass transfer from the compressed gas phase to the liquid phase. Analysis of the exhaust gas takes place at environmental pressure. That means that a simple correction to the overall system pressure in the respective bioreactor already provides a more rational way compared to a generic model to correct raw off-gas signal. Events like basic feed additions and resulting pH controller response to counteract pH shifts are perfectly detectable using the raw signal alone. The modelled pCO2 obviously underestimated the sample-based offline pCO2 after inoculation. Inoculation and dependent pH controller response lead to immediate changes in dissolved carbon dioxide levels, whereas headspace back mixing takes more time depending on aeration rate. Fast changes like base additions, set point changes or feed additions will have a delayed response in off-gas analysis. Relatively slow process changes in mammalian cell culture processes still allow for meaningful monitoring via exhaust carbon dioxide.
4. Discussion
Sample-based offline measurement of pH and pCO2 may not represent the true pH in the bioreactor with an accuracy that is required in the future. Many parameters potentially affect sample pH and pCO2 after sampling. In particular, pH is a parameter that has tremendous impact on process performance. Bioreactor probe signals must be considered a function of respective sample-based offline measurement after readjustment. Procedures to calibrate pH probes that are stated in respective Pharmacopeias or technical references issued by, e.g., the National Institute of Standards and Technology (NIST), are all based on certified pH buffers. Those buffers are designed to be as stable and as independent to environmental conditions like temperature as possible. However, this approach does not necessarily guarantee comparable results in samples that contain cells in cell culture media. Sample properties, sample hold times and local equipment and procedures may alter sample pH in different ways between sites, plants or even operators. Furthermore, buffers are commonly used for sensor calibration at temperatures other than sample temperature, which cannot be compensated for with typical automatic temperature compensation (ATC). Keeping sample temperatures constant is one way to achieve more comparable results. On the other hand reheating cooled samples to fermentation temperature includes longer sample hold times, again altering sample pH. A reference method to prove pH comparable between sites does not yet exist, because samples cannot be measured simultaneously at two sites. Furthermore, samples containing cells cannot be stored or shipped to measure pH somewhere else because effects of cells and the carbonate buffer system would unpredictably alter sample pH.
In this paper, we propose a method that is able to resolve pH with superior accuracy compared to sample-based offline measurement. Because pH can be determined without the need to sample, all potential offsets introduced by sample-based offline measurement are avoidable. Off-gas analysis thereby is relatively easy to establish in GMP manufacturing because measurement takes place outside the sterile barrier. The underlying fundamental chemical principal is very well described and can universally be applied.
The proposed method for pH determination is applicable in cell free media. After inoculation, exhaust carbon dioxide concentration becomes a sum parameter affected mainly by respiration, feed addition, lactate generation and pH controller response. After adjustment of bioreactor pH-probe signals based on the exhaust carbon dioxide concentration, further adjustments based on offline measurement should be avoided. This may question standard pH readjustment strategies based on frequent offline measurement as a detection measure for unintended bioreactor probe drifts.
How well a potential bioreactor probe drift is detectable with offline measurement heavily depends on changing sample properties during fermentation and the overall variability of a respective sample-based offline measurement. Variability directly affects the information content of a single data point derived by, e.g., daily sampling. The data shown alone shows that mean offsets may be small, but overall variability of sample-based offline measurement massively decreases efficacy to accurately determine the true pH in bioreactors.
For internal purposes, delta probe alarms were utilized to detect single probe drifts online in bioreactors that are equipped with more than one pH probe. Probability of an undetectable double probe drift thereby is considered low. Having efficient detectability measures in place to identify potential bioreactor probe drifts thereby reduces the dependency on sample-based offline measurements.
When pH can be determined by exhaust carbon dioxide concentration, pCO2 can be derived by the very same principles as well. The main advantage is that there is no need for balancing; carbonate concentrations in feed and media solutions as well as carbonate influx simply do not matter for deriving a continuous online signal that is superior to offline values. Data quality derived by pCO2 probes thereby may be questionable if either are calibrated by defined gas flow added by mass flow controllers or by sample-based offline readings. Accuracy of both methods is limited, leading to limited accuracy of pCO2 probe signals.
We presented this here together with pH to highlight that having to validate methods against a standard that is flawed by sampling and offline measurement proves difficult. Knowing that offsets are unavoidable for both pH and pCO2, strong rationales or scientific principles are needed to implement more accurate methods, even if they do not necessarily match the reference due to those offsets. We have to finally accept that a standard that obviously was sufficient for decades may not be sufficient in the future.
For scaling purposes, addressing carbon dioxide removal is extremely easy using off-gas analysis. Carbon dioxide removal equals gas flow times carbon dioxide exhaust concentration. This enables one to directly address aeration strategies without the need to know bioreactor configuration and the usual parameters like kLa, which are notoriously insufficient to match carbon dioxide removal and solubility between a small and large scale.
5. Conclusions
The true bioreactor pH cannot be accurately determined with sample-based offline measurements. Sample-based offline measurements may furthermore be efficient detection measures only for bioreactor pH-probe drifts that are relatively large.
Therefore, we recommend decreasing the dependency on sample-based offline measurements as far as possible. The proposed carbon dioxide-derived method represents a suitable alternative that is applicable in a variety of fermentation systems like stainless steel or single use bioreactors with acceptable effort. In fact, the method described here already has been successfully used for process transfers from a 2 L (Development) up to 12k scale (GMP Manufacturing); not one single sample-based offline pH value has been used to adjust bioreactor probes in the process.
If a sample-based offline measurement is used, we encourage authors to describe exactly how pH was measured including sample hold times, offline methods, calibration routines and readjustment strategies. Findings are not necessarily reproducible if the pH control strategy is unknown, making scientific progress in mammalian cell culture especially slow.
The ability to control pH more accurately will have tremendous impact on many industrial needs in the future. Bringing down process variability finally enables us to detect other effectors that are now undetectable and unquantifiable due to underlying process variability, and this variability is significantly driven by pH variability. The impact of raw materials, seed train and inoculum train operations, media preparation and so on will be better quantifiable.
More sophisticated approaches, such as complex modelling and soft sensoring, proteomics/transcriptomics, in silico approaches, and advanced analytics that also depend on pH, will only yield more relevant findings if data quality increases. Current pH variability in mammalian cell culture inhibits faster progress in all areas related to mammalian cell culture, and the potential of tighter and more accurate pH control cannot be underestimated.
6. Patent Applications
WO/2017/072340 MONITORING STATE DEVIATIONS IN BIOREACTORS, PCT/EP/2016/076167 Applicant F.HOFFMANN-LA ROCHE AG.
WO/2017/072346 IDENTIFICATION OF CALIBRATION DEVIATIONS OF PH-MEASURING DEVICES, PCT/EP/2016/076173 Applicant F.HOFFMANN-LA ROCHE AG.
Author Contributions
Conceptualization C.K., Methodology C.K., formal analysis T.W. for SUBs, V.T. for pCO2, C.K. for pH reference method writing—original draft preparation, C.K., V.T., T.W. writing—review and editing, C.K., V.T., T.W.; visualization, C.K., V.T., T.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work is exclusively funded by F. Hoffmann-La Roche AG.
Acknowledgments
This work is a small excerpt of a global development and implementation initiative within the Roche Drug Substance Network, where people of all functions including regulatory, procurement, manufacturing, development, research, quality functions and management drive and fund ongoing efforts. Eppendorf and BlueSens were especially supportive providing in-depth knowledge and device upgrades for GMP implementation.
Conflicts of Interest
Author Christian Klinger is also Inventor of the carbon dioxide-based pH reference method and therefore has a potential conflict of interest. F. Hoffmann-La Roche AG is funder and owner of the patent application “the carbon dioxide based pH reference method” and therefore has a potential conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. Certainly, the funders had particular interest in the outcome and benefits that may be realized by developing and implementing the proposed method.
References
- Michl, J.; Park, K.C.; Swietach, P. Evidence-based guidelines for controlling pH in mammalian live-cell culture systems. Commun. Biol. Vol. 2019, 2, 144. [Google Scholar] [CrossRef] [PubMed]
- de Pool, J.D.; Van Den Berg, S.A.; Pilgram, G.S.; Ballieux, B.E.; Van Der Westerlaken, L.A. Validation of the blood gas analyzer for pH measurements in IVF culture medium: Prevent suboptimal culture conditions. PLoS ONE 2018, 13, e0206707. [Google Scholar] [CrossRef] [PubMed]
- Frahm, B.; Blank, H.C.; Cornand, P.; Oelssner, W.; Guth, U.; Lane, P.; Munack, A.; Johannsen, K.; Pörtner, R. Determination of dissolved CO(2) concentration and CO(2) production rate of mammalian cell suspension culture based on off-gas measurement. J. Biotechnol. 2002, 99, 133–148. [Google Scholar] [CrossRef]
- Lawrence, J. Henderson. Concerning the relationship between the strength of acids and their capacity to preserve neutrality. Am. J. Physiol. 1908, 21, 173–179. [Google Scholar]
- Busse, C.; Biechele, P.; de Vries, I.; Reardon, K.F.; Solle, D.; Scheper, T. Sensors for disposable bioreactors. Eng. Life Sci. 2017, 17, 940–952. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).