A Reliable Automated Sampling System for On-Line and Real-Time Monitoring of CHO Cultures
Abstract
:1. Introduction
- Quantification of the automated liquid handling and analysis
- Optimization and adaptation of an HPLC method for amino acid measurement
- Application on a mammalian cell culture fed batch
- Comparison to other real-time monitoring systems
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. HPLC Methods
2.3. Biochemical Methods
2.4. Set-Up
2.5. The Automated Sampling System
2.6. Software and Data Management
2.7. Cultivations
2.8. Technical Run
3. Results
3.1. System Performance
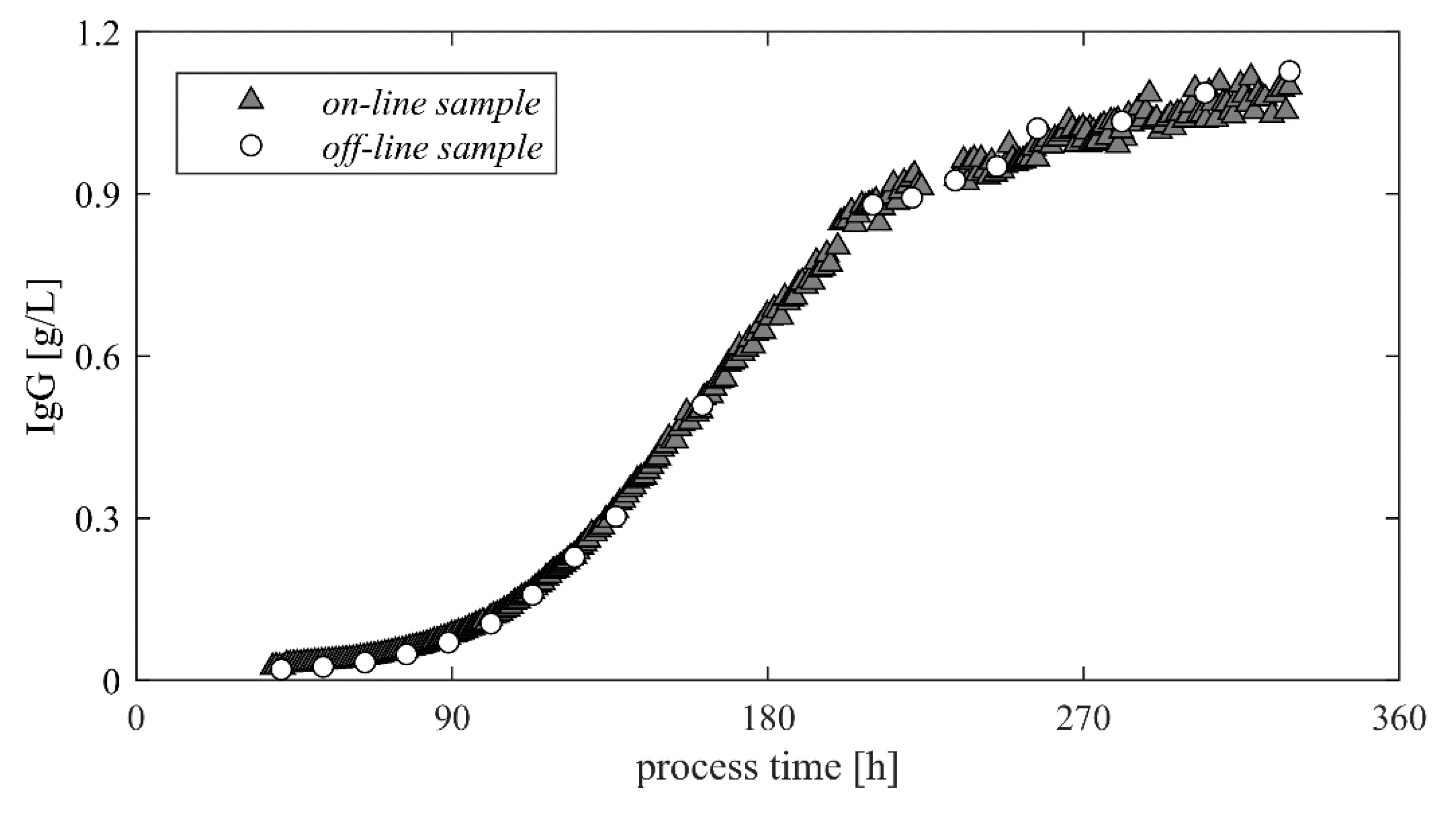
3.2. Monitoring—HPLC Autosampler Required
3.3. Monitoring—HPLC Direct Transfer
3.4. Monitoring—Bio HT Direct Transfer
4. Discussion
4.1. Automated Sampling in General
- (1)
- Does the system impact the sterility of the bioprocess?—No.
- (2)
- How much volume and how much dead volume are drawn from the bioreactor?—The sample volume is approx. 3.5 mL, of which about 1 mL is dead volume.
- (3)
- Does the system support sample processing (i.e., dilution, cell removal etc.)?—Yes.
- (4)
- Does the automated procedure impact the analytical result (i.e., dilution effects)?—There is a constant dilution factor observable, which can be included in final calculations.
- (5)
- How is the communication between process, sampling system, and analyzers realized?—The communication was achieved with one software (Lucullus PIMS) that coordinated the sample draw as well as the transfer of the sample to the analyzer and the initiation of the measurement.
4.2. Assessment of the Presented Liquid Handling System
- (1)
- The system was tested for CHO cells with a maximum viable cell density of about 12∙106 cells/mL, resulting in a required adaptation of the POT to reach the desired volume of permeate after filtration. Currently, the trend is towards high cell density cultivations in perfusion mode, which were not tested in this contribution. The actual filter area in the filtration module is 3.14 cm2, which is filtering 2.5 mL of cell suspension. An increase of the filter area might have a positive impact on the permeate volume.
- (2)
- During the operation of the system (especially when applying the Protein A HPLC method), it was observed that the precolumn had to be changed approximately 8–10 times during one cultivation run. The required changing frequency increased with progress of the process. This leads to the conclusion that the load of particles of the samples after the automated filtration is higher than after manual cell removal. A reason could be the filtration procedure per se or the applied pore size of the filter of 0.45 µm. Possible solutions to overcome this problem are (i) the use of a membrane with 0.22 µm pore size or (ii) the introduction of a prefiltration step or a dual filtration.
- (3)
- 1–2% of the samples resulted in empty vials after the filtration step.
4.3. Comparison with In-Line Sensors
4.4. Outlook
5. Conclusions
- Depending on the preprocessing and the applied analytical method, systematic deviations were observed. They were mainly caused by dilution effects and can be assumed to be constant. On the other hand, the random error seems to be significantly reduced.
- An existing HPLC method for amino acid analysis was successfully adapted in a way that it can now be applied for full automated on-line monitoring.
- The automated sampling and analytic system was successfully tested in mammalian cell culture fed-batch processes. The monitoring of various analytes was performed without significant errors or system failures. The higher measurement frequency and strongly reduced random errors resulted in a larger information content per experiment.
- The accuracy of the system is an order of magnitude better than the compared methods.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

| Sample | VCC [106 Cells/mL] | TCC [106 Cells/mL] | Viability [%] | Debris * [106 Cells/mL] |
|---|---|---|---|---|
| Manual 1 | 3.34 | 3.44 | 97.00 | 0.35 |
| Manual 2 | 3.45 | 3.69 | 93.00 | 0.48 |
| Manual 3 | 3.25 | 3.51 | 92.70 | 0.37 |
| Manual 4 | 3.40 | 3.62 | 94.00 | 0.42 |
| Manual 5 | 2.79 | 3.01 | 92.60 | 0.48 |
| Numera 1.1 | 3.68 | 3.91 | 94.00 | 0.47 |
| Numera 1.2 | 3.29 | 3.50 | 94.10 | 0.57 |
| Numera 2.1 | 3.27 | 3.55 | 92.30 | 0.43 |
| Numera 2.2 | 3.14 | 3.32 | 94.50 | 0.45 |
References
- Sonnleitner, B. Automated Measurement and Monitoring of Bioprocesses: Key Elements of the M 3 C Strategy. In Measurement, Monitoring, Modelling and Control of Bioprocesses; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–33. [Google Scholar]
- Claßen, J.; Aupert, F.; Reardon, K.F.; Solle, D.; Scheper, T. Spectroscopic sensors for in-line bioprocess monitoring in research and pharmaceutical industrial application. Anal. Bioanal. Chem. 2017, 409, 651–666. [Google Scholar] [CrossRef] [PubMed]
- Chong, L.; Saghafi, M.; Knappe, C.; Steigmiller, S.; Matanguihan, C.; Goudar, C.T. Robust on-line sampling and analysis during long-term perfusion cultivation of mammalian cells. J. Biotechnol. 2013, 165, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Kroll, P.; Sagmeister, P.; Reichelt, W.; Neutsch, L.; Klein, T.; Herwig, C. Ex situ online monitoring: Application, challenges and opportunities for biopharmaceuticals processes. Pharm. Bioprocess. 2014, 2, 285–300. [Google Scholar] [CrossRef]
- Cimander, C.; Bachinger, T.; Mandenius, C.-F. Integration of distributed multi-analyzer monitoring and control in bioprocessing based on a real-time expert system. J. Biotechnol. 2003, 103, 237–248. [Google Scholar] [CrossRef]
- Foley, D.A.; Wang, J.; Maranzano, B.; Zell, M.T.; Marquez, B.L.; Xiang, Y.; Reid, G.L. Online NMR and HPLC as a reaction monitoring platform for pharmaceutical process development. Anal. Chem. 2013, 85, 8928–8932. [Google Scholar] [CrossRef]
- Spadiut, O.; Dietzsch, C.; Posch, A.; Herwig, C. Evaluating online sampling probes for substrate concentration and protein production by a Design of Experiments screening approach. Eng. Life Sci. 2012, 12, 507–513. [Google Scholar] [CrossRef]
- US FDA. Guidance for industry PAT-A Framework for Innovative Pharmaceutical Development, Manufacturing, and Quality Assurance. Available online: www.fda.gov/downloads/Drugs/Guidances/ucm070305.pdf (accessed on 29 April 2020).
- Duarte, T.M.; Carinhas, N.; Barreiro, L.C.; Carrondo, M.J.; Alves, P.M.; Teixeira, A.P. Metabolic responses of CHO cells to limitation of key amino acids. Biotechnol. Bioeng. 2014, 111, 2095–2106. [Google Scholar] [CrossRef]
- Crowell, C.K.; Grampp, G.E.; Rogers, G.N.; Miller, J.; Scheinman, R.I. Amino acid and manganese supplementation modulates the glycosylation state of erythropoietin in a CHO culture system. Biotechnol. Bioeng. 2007, 96, 538–549. [Google Scholar] [CrossRef]
- Zalai, D.; Hevér, H.; Lovász, K.; Molnár, D.; Wechselberger, P.; Hofer, A.; Párta, L.; Putics, Á.; Herwig, C. A control strategy to investigate the relationship between specific productivity and high-mannose glycoforms in CHO cells. Appl. Microbiol. Biotechnol. 2016, 100, 7011–7024. [Google Scholar] [CrossRef] [Green Version]
- Hofer, A.; Herwig, C. Quantitative determination of nine water-soluble vitamins in the complex matrix of corn steep liquor for raw material quality assessment. J. Chem. Technol. Biotechnol. 2017, 92, 2106–2113. [Google Scholar] [CrossRef]
- Hofer, A.; Hauer, S.; Kroll, P.; Fricke, J.; Herwig, C. In-depth characterization of the raw material corn steep liquor and its bioavailability in bioprocesses of Penicillium chrysogenum. Process Biochem. 2018, 70, 20–28. [Google Scholar] [CrossRef]
- Bhatia, H.; Mehdizadeh, H.; Drapeau, D.; Yoon, S. In-line monitoring of amino acids in mammalian cell cultures using raman spectroscopy and multivariate chemometrics models. Eng. Life Sci. 2018, 18, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Larson, T.M.; Gawlitzek, M.; Evans, H.; Albers, U.; Cacia, J. Chemometric evaluation of on-line high-pressure liquid chromatography in mammalian cell cultures: Analysis of amino acids and glucose. Biotechnol. Bioeng. 2002, 77, 553–563. [Google Scholar] [CrossRef]
- Cimander, C.; Mandenius, C.-F. Online monitoring of a bioprocess based on a multi-analyser system and multivariate statistical process modelling. J. Chem. Technol. Biotechnol. 2002, 77, 1157–1168. [Google Scholar] [CrossRef]
- Van de Merbel, N.C.; Lingeman, H.; Brinkman, U.A. Sampling and analytical strategies in on-line bioprocess monitoring and control. J. Chromatogr. A 1996, 725, 13–27. [Google Scholar] [CrossRef]
- Esmonde-White, K.A.; Cuellar, M.; Uerpmann, C.; Lenain, B.; Lewis, I.R. Raman spectroscopy as a process analytical technology for pharmaceutical manufacturing and bioprocessing. Anal. Bioanal. Chem. 2017, 409, 637–649. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.L.T.; Boccazzi, P.; Gorret, N.; Ram, R.J.; Sinskey, A.J. In situ bioprocess monitoring of Escherichia coli bioreactions using Raman spectroscopy. Vib. Spectrosc. 2004, 35, 131–137. [Google Scholar] [CrossRef]
- Müller, J.; Knop, K.; Wirges, M.; Kleinebudde, P. Validation of Raman spectroscopic procedures in agreement with ICH guideline Q2 with considering the transfer to real time monitoring of an active coating process. J. Pharm. Biomed. Anal. 2010, 53, 884–894. [Google Scholar] [CrossRef]
- Borys, M.C.; Linzer, D.I.H.; Papoutsakis, E.T. Culture pH affects expression rates and glycosylation of recombinant mouse placental lactogen proteins by Chinese hamster ovary (CHO) cells. Biotechnology 1993, 11, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Lee, G.M. Differences in optimal pH and temperature for cell growth and antibody production between two Chinese hamster ovary clones derived from the same parental clone. J. Microbiol. Biotechnol. 2007, 17, 712–720. [Google Scholar] [PubMed]
- Yoon, S.K.; Choi, S.L.; Song, J.Y.; Lee, G.M. Effect of culture pH on erythropoietin production by Chinese hamster ovary cells grown in suspension at 32.5 and 37.0 °C. Biotechnol. Bioeng. 2005, 89, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Franco, R.; Daniela, G.; Fabrizio, M.; Ilaria, G.; Detlev, H. Influence of osmolarity and pH increase to achieve a reduction of monoclonal antibodies aggregates in a production process. Cytotechnology 1999, 29, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Birch, J.R.; Racher, A.J. Antibody production. Adv. Drug. Deliv. 2006, 58, 671–685. [Google Scholar] [CrossRef] [PubMed]
- Kroll, P.; Stelzer, I.V.; Herwig, C. Soft sensor for monitoring biomass subpopulations in mammalian cell culture processes. Biotechnol. Lett. 2017, 39, 1667–1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| Correlation | Slope | Offset | Confidence Interval Slope (95%) | R2 |
|---|---|---|---|---|
| Off-line vs on-line | 0.8409 | 0.0056 | 0.8216–0.8602 | 0.9981 |
| Off-line vs on-line calibrated via Numera | 0.9696 | 0.0133 | 0.9474–0.9918 | 0.9981 |
| Off-line vs on-line considering DF | 0.9973 | 0.0067 | 0.9745–1.0202 | 0.9981 |
| Reference Method (HPLC) | Automated Sampling and On-Line HPLC | Raman Spectroscopy | |
|---|---|---|---|
| cv(RMSE) [-] | cv(RMSE) [-] | cv(RMSE) [-] | |
| Tyrosine | 0.012 | 0.058 | 0.53 * |
| Tryptophan | 0.020 | 0.066 | 0.27 * |
| Phenylalanine | 0.016 | 0.062 | 0.47 * |
| Methionine | 0.025 | 0.071 | 0.29 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hofer, A.; Kroll, P.; Barmettler, M.; Herwig, C. A Reliable Automated Sampling System for On-Line and Real-Time Monitoring of CHO Cultures. Processes 2020, 8, 637. https://doi.org/10.3390/pr8060637
Hofer A, Kroll P, Barmettler M, Herwig C. A Reliable Automated Sampling System for On-Line and Real-Time Monitoring of CHO Cultures. Processes. 2020; 8(6):637. https://doi.org/10.3390/pr8060637
Chicago/Turabian StyleHofer, Alexandra, Paul Kroll, Matthias Barmettler, and Christoph Herwig. 2020. "A Reliable Automated Sampling System for On-Line and Real-Time Monitoring of CHO Cultures" Processes 8, no. 6: 637. https://doi.org/10.3390/pr8060637
APA StyleHofer, A., Kroll, P., Barmettler, M., & Herwig, C. (2020). A Reliable Automated Sampling System for On-Line and Real-Time Monitoring of CHO Cultures. Processes, 8(6), 637. https://doi.org/10.3390/pr8060637






