Time-Course Transcriptome Analysis Reveals Global Gene Expression Profiling and Dynamic Developmental Signatures across Complete Life Cycle of Bombyx mori
Abstract
:1. Introduction
2. Materials and Methods
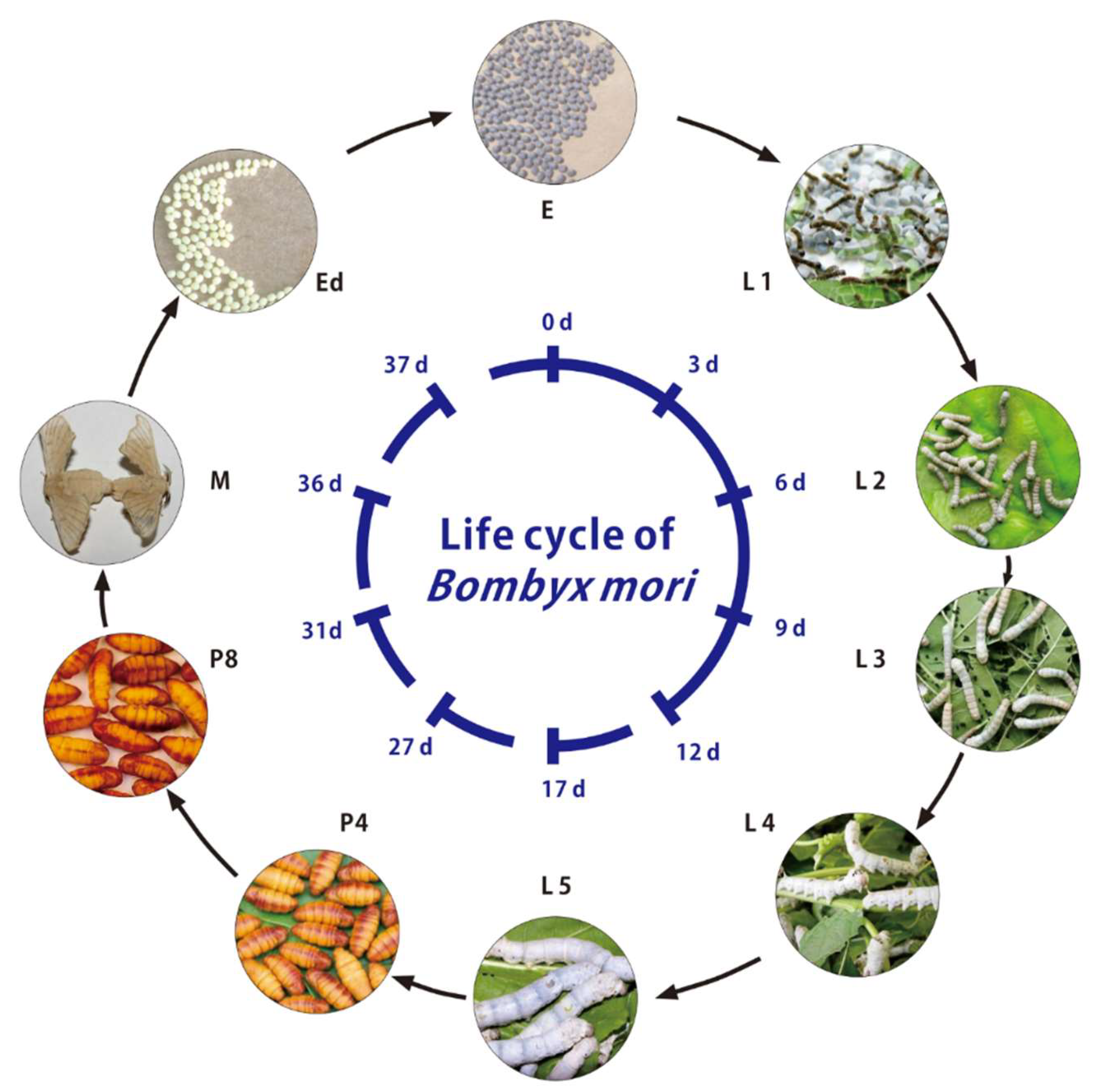
2.1. Silkworm Strain and Sample Preparation
2.2. RNA Extraction, cDNA Library Construction, and RNA-Sequencing
2.3. Bioinformatics Analysis
3. Results
3.1. Experimental Design and Overview of the Transcriptome Sequencing Data
3.2. Gene Expression Profile Dynamics throughout Silkworm Complete Life Cycle
3.3. Gene Expression Profiles during Embryogenesis
3.4. Gene Expression Differences throughout Silkworm Larval Development Stage
3.5. Gene Expression Differences throughout Silkworm Adult Development Stages
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ye, C.; Jiang, S.; Gong, M.; Min, Q.; Fan, M.; Gao, J.; Meng, Y. Expressional Localization and Functionally Identifying an RNA Editing Enzyme BmADARa of the Silkworm Bombyx mori. Insects 2020, 11, 523. [Google Scholar] [CrossRef] [PubMed]
- Brady, D.; Saviane, A.; Cappellozza, S.; Sandrelli, F. An Efficient Workflow for Screening and Stabilizing CRISPR/Cas9-Mediated Mutant Lines in Bombyx mori. Methods Protoc. 2020, 4, 4. [Google Scholar] [CrossRef]
- Tabunoki, H.; Bono, H.; Ito, K.; Yokoyama, T. Can the silkworm (Bombyx mori) be used as a human disease model? Drug Discov. Ther. 2016, 10, 3–8. [Google Scholar] [CrossRef]
- Abdelli, N.; Peng, L.; Keping, C. Silkworm, Bombyx mori, as an alternative model organism in toxicological research. Environ. Sci. Pollut. Res. Int. 2018, 25, 35048–35054. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Fang, Y.; Li, D.C.; Chen, D.S.; Wu, F. Effect of Lactic Acid Supplementation on the Growth and Reproduction of Bombyx mori (Lepidopteria: Bombycidae). J. Insect Sci. 2021, 21, 7. [Google Scholar] [CrossRef]
- Xu, G.; Yi, Y.; Lyu, H.; Gong, C.; Feng, Q.; Song, Q.; Peng, X.; Liu, L.; Zheng, S. DNA methylation suppresses chitin degradation and promotes the wing development by inhibiting Bmara-mediated chitinase expression in the silkworm, Bombyx mori. Epigenetics Chromatin 2020, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Xu, H.Q.; Chen, D.; Zhang, S.Y.; Li, W.J.; Smagghe, G.; Wang, J.J. Genome-wide gene expression profiling of the melon fly, Zeugodacus cucurbitae, during thirteen life stages. Sci. Data 2020, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Bellone, R.; Failloux, A.B. The Role of Temperature in Shaping Mosquito-Borne Viruses Transmission. Front. Microbiol. 2020, 11, 584846. [Google Scholar] [CrossRef]
- Li, H.M.; Yang, B.Z.; Zhang, X.J.; Jiang, H.Y.; Li, L.M.; Ahmad, H.I.; Chen, J.P. Transcriptome analysis reveals the genetic basis underlying the development of skin appendages and immunity in hedgehog (Atelerix albiventris). Sci. Rep. 2020, 10, 13920. [Google Scholar] [CrossRef]
- Wu, M.; Wang, P.; Gao, M.; Shen, D.; Zhao, Q. Transcriptome analysis of the eggs of the silkworm pale red egg (rep-1) mutant at 36 hours after oviposition. PLoS ONE 2020, 15, e0237242. [Google Scholar] [CrossRef]
- Gong, J.; Zheng, X.; Zhao, S.; Yang, L.; Xue, Z.; Fan, Z.; Tang, M. Early Molecular Events during Onset of Diapause in Silkworm Eggs Revealed by Transcriptome Analysis. Int. J. Mol. Sci. 2020, 21, 6180. [Google Scholar] [CrossRef]
- Xu, P.Z.; Zhang, M.R.; Wang, X.Y.; Wu, Y.C. Precocious Metamorphosis of Silkworm Larvae Infected by BmNPV in the Latter Half of the Fifth Instar. Front. Physiol. 2021, 12, 650972. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhu, F.; Xiao, R.; Ge, Q.; Tang, H.; Kong, M.; Taha, R.H.; Chen, K. Increased expression of Suppressor of cytokine signaling 2 (BmSOCS2) is correlated with suppression of Bombyx mori nucleopolyhedrovirus replication in silkworm larval tissues and cells. J. Invertebr. Pathol. 2020, 174, 107419. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Yuan, Y.; Zhu, F.; He, S.; Ge, Q.; Wang, X.; Taha, R.; Chen, K. Transcriptomics and proteomics-based analysis of heterosis on main economic traits of silkworm, Bombyx mori. J. Proteom. 2020, 229, 103941. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cheng, T.; Hu, W.; Peng, Z.; Liu, C.; Xia, Q. Genome-wide identification and analysis of JHBP-domain family members in the silkworm Bombyx mori. Mol. Genet. Genom. 2016, 291, 2159–2171. [Google Scholar] [CrossRef]
- Meng, X.; Zhu, F.; Chen, K. Silkworm: A Promising Model Organism in Life Science. J. Insect Sci. 2017, 17, 97. [Google Scholar] [CrossRef]
- Chen, A.; Li, Q.; Liao, P.; Zhao, Q.; Tang, S.; Wang, P.; Meng, G.; Dong, Z. Semaphorin-1a-like gene plays an important role in the embryonic development of silkworm, Bombyx mori. PLoS ONE 2020, 15, e0240193. [Google Scholar] [CrossRef]
- Deng, M.J.; Lin, X.D.; Wen, C.W.; Dong, M.J.; Lin, Q.T.; Zhang, S.Z.; Xu, J.P. Metabolic changes in the midgut of Eri silkworm after Oral administration of 1-deoxynojirimycin: A 1H-NMR-based metabonomic study. PLoS ONE 2017, 12, e0173213. [Google Scholar] [CrossRef]
- Fometu, S.S.; Wu, G.; Ma, L.; Davids, J.S. A review on the biological effects of nanomaterials on silkworm (Bombyx mori). Beilstein J. Nanotechnol. 2021, 12, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Bayega, A.; Djambazian, H.; Tsoumani, K.T.; Gregoriou, M.E.; Sagri, E.; Drosopoulou, E.; Mavragani-Tsipidou, P.; Giorda, K.; Tsiamis, G.; Bourtzis, K.; et al. De novo assembly of the olive fruit fly (Bactrocera oleae) genome with linked-reads and long-read technologies minimizes gaps and provides exceptional Y chromosome assembly. BMC Genom. 2020, 21, 259. [Google Scholar] [CrossRef]
- Suetsugu, Y.; Futahashi, R.; Kanamori, H.; Kadono-Okuda, K.; Sasanuma, S.; Narukawa, J.; Ajimura, M.; Jouraku, A.; Namiki, N.; Shimomura, M.; et al. Large scale full-length cDNA sequencing reveals a unique genomic landscape in a lepidopteran model insect, Bombyx mori. G3 (Bethesda) 2013, 3, 1481–1492. [Google Scholar] [CrossRef] [PubMed]
- Seib, F.P. Emerging Silk Material Trends: Repurposing, Phase Separation and Solution-Based Designs. Materials (Basel) 2021, 14, 1160. [Google Scholar] [CrossRef]
- Chen, Q.; Ma, Z.; Wang, X.; Li, Z.; Zhang, Y.; Ma, S.; Zhao, P.; Xia, Q. Comparative proteomic analysis of silkworm fat body after knocking out fibroin heavy chain gene: A novel insight into cross-talk between tissues. Funct. Integr. Genom. 2015, 15, 611–637. [Google Scholar] [CrossRef] [PubMed]
- Rehman, N.; Varghese, J. Larval nutrition influences adult fat stores and starvation resistance in Drosophila. PLoS ONE 2021, 16, e0247175. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Zhang, Y.; Ma, C.; Chen, H.; Guo, J.; Zhou, Z. Silencing the Myosin Regulatory Light Chain Gene sqh Reduces Cold Hardiness in Ophraella communa LeSage (Coleoptera: Chrysomelidae). Insects 2020, 11, 844. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Fan, Y.; Ge, Q.; Xu, J.; Taha, R.H.; Yuan, Y.; Chen, K. Time-Course Transcriptome Analysis Reveals Global Gene Expression Profiling and Dynamic Developmental Signatures across Complete Life Cycle of Bombyx mori. Processes 2021, 9, 1730. https://doi.org/10.3390/pr9101730
Wang X, Fan Y, Ge Q, Xu J, Taha RH, Yuan Y, Chen K. Time-Course Transcriptome Analysis Reveals Global Gene Expression Profiling and Dynamic Developmental Signatures across Complete Life Cycle of Bombyx mori. Processes. 2021; 9(10):1730. https://doi.org/10.3390/pr9101730
Chicago/Turabian StyleWang, Xueqi, Yixuan Fan, Qi Ge, Jia Xu, Rehab Hosny Taha, Yi Yuan, and Keping Chen. 2021. "Time-Course Transcriptome Analysis Reveals Global Gene Expression Profiling and Dynamic Developmental Signatures across Complete Life Cycle of Bombyx mori" Processes 9, no. 10: 1730. https://doi.org/10.3390/pr9101730





