Abstract
Ilex cornuta Leaves (ICLs) are a representative and traditional prescription for controlling obesity. Nevertheless, the corresponding therapeutic compounds and related pharmacological mechanisms of such medication remain undocumented. The compounds from ICLs were identified by gas chromatography-mass spectrum (GC-MS), and SwissADME confirmed their physicochemical properties. Next, the target proteins related to compounds or obesity-associated proteins were retrieved from public databases. RPackage constructed the protein–protein interaction (PPI) network, a bubble chart, and signaling pathways–target proteins–compounds (STC) network. Lastly, a molecular docking test (MDT) was performed to evaluate the affinity between target proteins and ligands from ICLs. GC-MS detected a total of 51 compounds from ICLs. The public databases identified 219 target proteins associated with selective compounds, 3028 obesity-related target proteins, and 118 overlapping target proteins. Moreover, the STC network revealed 42 target proteins, 22 signaling pathways, and 39 compounds, which were viewed to be remedially significant. The NOD-like receptor (NLR) signaling pathway was considered a key signaling pathway from the bubble chart. In parallel, the MDT identified three target proteins (IL6, MAPK1, and CASP1) on the NLR signaling pathway and four compounds against obesity. Overall, four compounds from ICLs might show anti-obesity synergistic efficacy by inactivating the NLR signaling pathway.
1. Introduction
Obesity has now sharply hit epidemic levels and has become a significant cause of global death [1]. A recent report indicates that obesity is closely linked to metabolic disorders that distinctly develop psychological stress and often exacerbate obesity-related complications [2]. Obesity can present in all ages; in 2016, almost 13% of the world population were overweight [3]. Moreover, obesity is deeply associated with other metabolic diseases such as diabetes, hypertension, atherosclerosis, and heart failure [4]. The main driving factors of metabolic disorders are cytokines, which are mainly implicated with a high-fat diet [5]. Currently, available anti-obesity medications include sibutramine, rimonabant, and orlistat, which may lead to side effects like diarrhea, fecal incontinence, flatulence, and dyspepsia [6,7]. In contrast, herbal solutions based on natural organic compounds have been used for thousands of years with high efficacy and safety [8]. To date, many natural herbal plants are used in a diet regime or as an alleviator for anti-obesity [9], for example, Ilex cornuta Leaves (ICLs) are potentially used to treat obesity. A topical patent on ICLs summarized that the extract could be effective for various metabolic diseases [10]. Moreover, some reports demonstrated that ICLs extract has potent anti-inflammatory effects associated with obesity in adipocytes [11,12]. Another study stated that the Ilex species, including ICLs, are known for regulating lipid metabolism and weight-loss activity [13]. Until now, research of ICLs has been focused on a broad range of metabolic disorders without defining the exact action mechanism for particular diseases. Therefore, the studies on active compounds and mechanisms of ICLs against obesity should be proven to understand the pharmacological value in alleviating obesity.
Network pharmacology is an efficient method to comprehend relationships between multiple unspecific compounds and multiple target proteins [14]. It is a relatively effective technique to analyze herbal medicines regarding novel active compounds and new mechanisms of action against different diseases [15]. Mainly, for metabolic syndrome research, the network pharmacology approach contributes to unravelling complex biological systems and interactions between active compounds and target proteins [16,17].
The concept and principle of network pharmacology proposed by Andrew Hopkins are based on bioinformatics and system biology [18]. In addition, the development of bioinformatics with system biology supports the progression of network pharmacology; for instance, Dr. Shoichet’s group expanded the ‘similarity ensemble approach’ to search the connectivity between ligand and target protein [19]. SwissTargetPrediction (STP) is a web-based bioinformatics database developed by the SIB (Swiss Institute of Bioinformatics), which loaded 376,342 reliable experimental compounds and 3,068 target proteins since 2014 [20]. Likewise, Dr. Furlong developed ‘DisGeNET’, a comprehensive platform for the efficient exploration of 380,000 associations between 13,000 diseases and more than 16,000 genes [21]. Furthermore, Online Mendelian Inheritance in Man (OMIM) is integrated with other genetic databases such as PubMed references, DNA and protein sequence, and the specific mutation database [22]. Thus, this study utilized these four databases to understand the relationships between compounds from ICLs and obesity-related target proteins from a human. To sum things up, network pharmacology utilized via four bioinformatics explored the mechanisms of ICLs against obesity and found promising active compounds from ICLs and associated target proteins. The functional diagram is exhibited in Figure 1.

Figure 1.
Workflow diagram of network pharmacology analysis of ICLs against obesity.
2. Results
2.1. Chemical Compounds from ICLs
A total of 52 chemical compounds in ICLs were identified by the GC-MS analysis (Figure 2), and the name of compounds, PubChem ID, retention time (min), peak area (%), and pharmacological activities were enlisted in Table 1. Lipinski’s rules accepted the number of 51 out of 52 chemical compounds (molecular weight ≤ 500g/mol; Moriguchi octanol-water partition coefficient ≤4.15; number of nitrogen or oxygen ≤10; number of NH or OH ≤5), and the selected 51 chemical compounds (excluding lactose) corresponded with the standard of ‘Abbott Bioavailability Score (>0.1)’ through SwissADME. Additionally, lactose was excluded due to the number of nitrogen or oxygen and the number of NH or OH. The TPSA (topological polar surface area) value of the selected 51 chemical compounds (excluding lactose) was also accepted (Table 2).
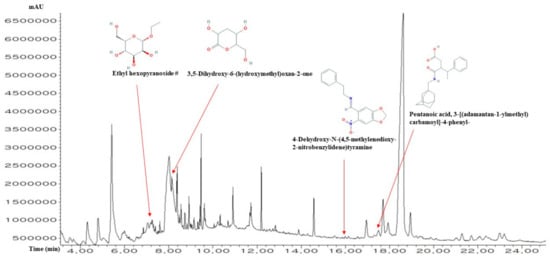
Figure 2.
A typical GC-MS peak of ICLs.

Table 1.
A list of 52 chemical compounds identified from ICLs via GC-MS and profiling of biological activities.

Table 2.
Physicochemical properties of chemical compounds for good oral bioavailability and cell membrane permeability.
2.2. Overlapping Target Proteins between SEA and STP
A total of 525 target proteins from SEA and 576 target proteins from STP connected to 51 chemical compounds were identified (Supplementary Table S1). The Venn diagram showed that 219 target proteins were overlapped between the two compound databases (Supplementary Table S1) (Figure 3A).
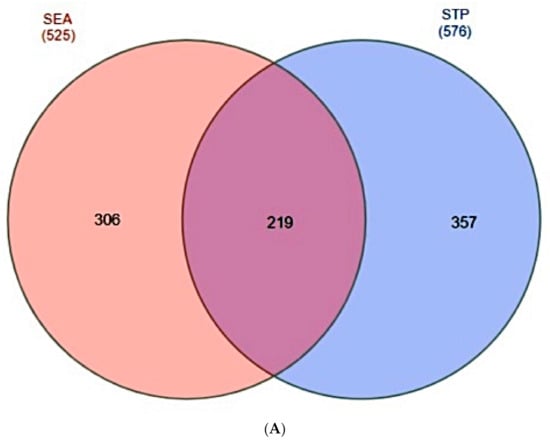

Figure 3.
(A) The overlapping targets (219) between SEA and STP databases. (B) The final targets (118) between the overlapping target proteins (OTPs) (219) and obesity-related target proteins (ORTPs) (3028).
2.3. Overlapping Target Proteins between Obesity-Related Target Proteins and 219 Target Proteins
A total of 3028 target proteins associated with obesity were selected by retrieval from DisGeNET and OMIM databases (Supplementary Table S2). The Venn diagram result revealed 118 overlapping target proteins between obesity-associated 3028 target proteins and the 219 overlapping target proteins (Figure 3B) (Supplementary Table S3).
2.4. PPI Networks
From STRING analysis, the final overlapping 118 target proteins were directly related to obesity occurrence and progression, indicating 116 nodes and 674 edges (Figure 4). Two (PAM and RGS4) out of 118 targets did not interact with any other targets. In PPI networks, IL6 manifested the highest degree (52), followed by VEGFA (47), PTGS2 (42), MAPK1 (36), and CASP3 (36) (Table 3). Hence, IL6 was considered the uppermost target protein in PPI networks.
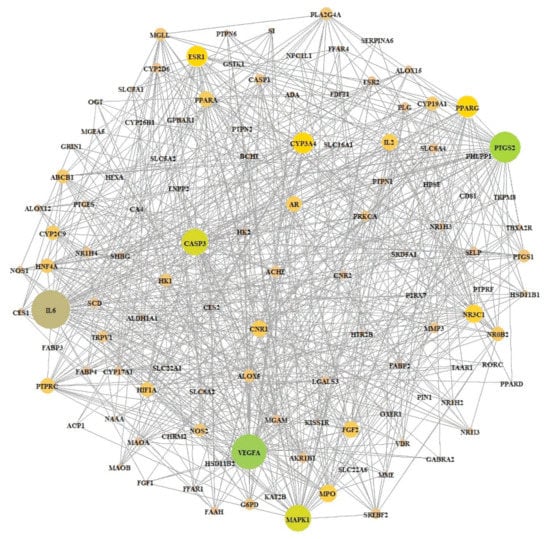
Figure 4.
PPI networks (116 nodes and 674 edges). The size of circles stands for the degree of values.

Table 3.
The degree value of 116 targets.
2.5. Analysis of Signaling Pathways against Obesity
The results of the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis revealed that 22 signaling pathways were related to 42 target proteins (false discovery rate < 0.05). The 22 signaling pathways were directly connected to obesity, suggesting that these 22 signaling pathways might be the significant pathways of ICLs against obesity. The description of 22 signaling pathways is provided in Table 4. A bubble chart showed that both NOD-like receptor signaling pathway and MAPK signaling pathway have the same rich factor of 0.024 (Figure 5). Additionally, NOD-like receptor signaling pathway was directly related to IL6 (the highest degree of value) but MAPK signaling pathway was unconnected to IL6 (listed in Table 4).

Table 4.
The number of 42 target proteins in 22 signaling pathways enrichment associated with obesity.

Figure 5.
Bubble chart of ICLs against obesity.
2.6. STC Networks Analysis of ICLs against Obesity
STC networks of ICLs against obesity are exhibited in Figure 6. There were 22 pathways, 42 target proteins, and 39 compounds (103 nodes, 333 edges). The nodes stand for a total number of three components: signaling pathways—target proteins—compounds (STC). The edges stand for the association of a total number of three components. The STC networks suggested that the network was associated with the therapeutic efficacy against obesity. Particularly, MAPK1 target protein (the highest degree value) was related to 20 out of 22 signaling pathways in STC networks, connected to NOD-like receptor signaling pathway. The main three target proteins of ICLs against obesity are indicated in the KEGG pathway diagram (Figure 7).
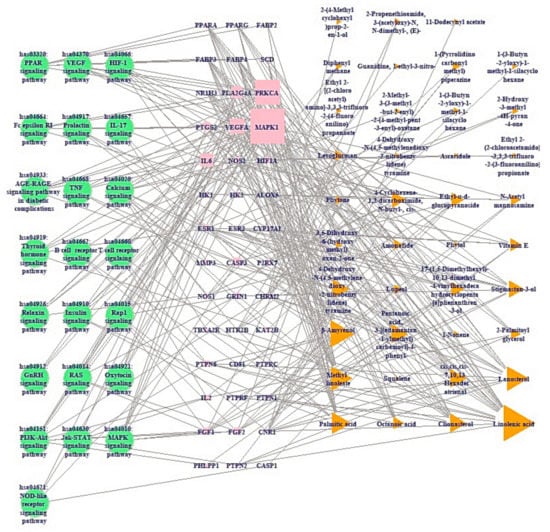
Figure 6.
STC networks. Green circle: signaling pathway; pink square: target protein; orange triangle: compound.
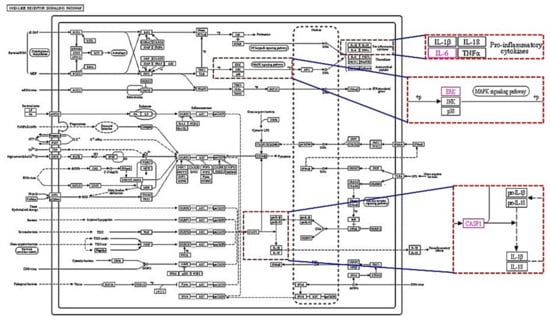
Figure 7.
NOD-like receptor signaling pathway. Red dotted line indicated IL6; MAPK1 (known as ERK); and CASP1 location. Pink colors represent target proteins of ICLs on obesity.
2.7. KEGG Pathway Analysis of NLR Signaling Pathway
KEGG pathway revealed location of the three target proteins (IL6, MAPK1, and CASP1) in intracellular components (Figure 7).
2.8. MDT of Four Target Proteins and 11 Compounds Connected to NLR Signaling Pathway
The IL6 protein (PDB ID: 4NI9) connected to three compounds on the NLR signaling pathway was used to perform MDT. It was observed that 3,5-dihydroxy-6-(hydroxymethyl)oxan-2-one) docked on the IL6 protein (PDB ID: 4NI9) exhibited the highest binding energy (−6.3 kcal/mol), followed by ethyl- α-d-glucopyranoside (−6.1 kcal/mol) and linolenic acid (−4.1 kcal/mol). The 3,5-dihydroxy-6-(hydroxymethyl)oxan-2-one also had the higher affinity than a positive control (veratric acid) [27] which showed −6.1 kcal/mol. The docking details of three compounds and a positive control are shown in Figure 8A, Table 5. The MDT of three compounds on MAPK1 (PDB ID: 4IZ5) was analyzed to identify the affinity. It was uncovered that 4-dehydroxy-N-(4,5-methylenedioxy-2-nitrobenzylidene)tyramine docked on an MAPK1 protein (PDB ID: 4IZ5) exposed the highest binding energy (−7.0 kcal/mol), followed by linolenic acid (-4.6 kcal/mol) and palmitic acid (−4.4 kcal/mol). Noticeably, 4-dehydroxy-N-(4,5-methylenedioxy-2-nitrobenzylidene)tyramine demonstrated higher affinity than a positive control (CU-Cpt 22) [28] showed −6.0 kcal/mol. The docking details of three compounds and a positive control are shown in Figure 8B, Table 6. The MDT score of four compounds on a P2RX7 protein (PDB ID: 5U2H) was analyzed to identify the affinity. It was exposed that pentanoic acid; 3-[(adamantan-1-ylmethyl)carbamoyl]-4-phenyl- docked on a P2RX7 protein (PDB ID: 5U2H) demonstrated the highest binding energy (−5.9 kcal/mol), followed by 4-cyclohexene-1,2-dicarboximide, N-butyl-, cis- (−4.1 kcal/mol), phytone (−3.3 kcal/mol), and cis,cis,cis-7,10,13-hexadecatrienal (−3.1 kcal/mol). The docking details of four compounds are shown in Table 7. Collectively, IL6 (PDB ID: 4NI9), MAPK1 (PDB ID:4IZ5), and P2RX7 (PDB ID: 5U2H) indicated that the affinity of each compound did not give a valid binding score (>−6.0 kcal/mol) [29]. Moreover, the affinity of four compounds on P2RX7 (PDB ID: 5U2H) was lower than the positive control (KN-62) [30] showed −9.8 kcal/mol.
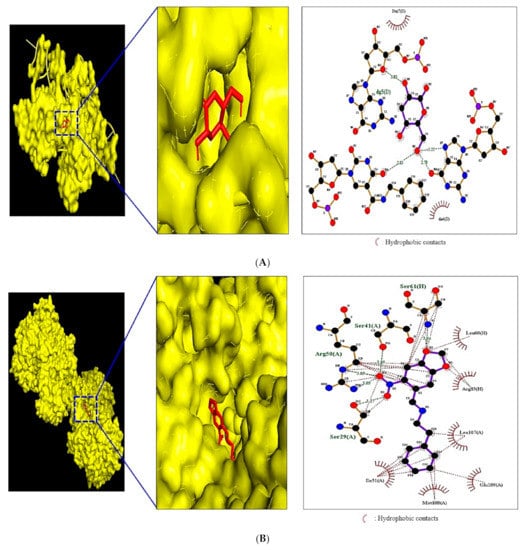

Figure 8.
(A) MDT of three, 3,5-Dihydroxy-6-(hydroxymethyl) oxan-2-one on IL6 (PDB ID: 4NI9). (B) MDT of 4-dehydroxy-N-(4, 5-methylenedioxy- 2- nitrobenzylidene) tyramine on MAPK1 (PDB ID: 4IZ5). (C) MDT of pentanoic acid, 3-[(adamantan-1-ylmethyl) carbamoyl]-4-phenyl- on CASP1 (PDB ID: 3D6F).

Table 5.
The binding energy of potential compounds and a positive control on IL6 (PDB ID: 4NI9).

Table 6.
Binding energy of potential compounds and a positive control on MAPK1 (PDB ID: 4IZ5).

Table 7.
Binding energy of potential compounds and a positive control on P2RX7 (PDB ID: 5U2H).
In addition, pentanoic acid; 3-[(adamantan-1-ylmethyl)carbamoyl]-4-phenyl-) docked on a CASP1 protein (PDB ID: 3D6F) showed a valid affinity (−7.3 kcal/mol), which was a comparatively higher affinity than three positive standard ligands (belnacasan [31], mulberroside A [32], and Q-VD-Oph [33]). The docking details of three compounds and the positive control are shown in Figure 8C, Table 8.

Table 8.
The binding energy of potential compounds and three positive controls on CASP1 (PDB ID: 3D6F).
2.9. Toxicological Properties of Four Key Compounds
Additionally, toxicological properties of ethyl- α-d-glucopyranoside; 3,5-dihydroxy-6-(hydroxymethyl)oxan-2-one; 4-dehydroxy-N-(4,5-methylenedioxy-2-nitrobenzylidene)tyramine; and pentanoic acid, 3-[(adamantan-1-ylmethyl)carbamoyl]-4-phenyl- were predicted by admetSAR online tool. Our result suggested that the bioactives did not relate to Ames toxicity, carcinogenic properties, acute oral toxicity, and rat acute toxicity properties (Table 9).

Table 9.
Toxicological properties of the highest affinity ligands on NOD-like receptor signaling pathway.
3. Discussion
PPI networks indicated that IL6 was the uppermost target protein (based on the highest degree of value: 52 degrees) to treat obesity. Another STC network suggested that the therapeutic efficacy of ICLs on obesity was directly associated with 22 signaling pathways, 42 target proteins, and 39 compounds. The network exposed that mitogen-activated protein kinase 1 (MAPK1) (known as ERK) with the highest degree ranking (20 degrees), was the most significant target protein of ICLs against obesity. In this analysis, the NLR signaling pathway was directly related to both IL6 and MAPK1, whereas the MAPK signaling pathway was not connected to IL6.
Thereby, the NLR signaling pathway was considered as a hub-signaling pathway of ICLs to ameliorate obesity; by constructing and analyzing the STC network, nine key compounds and four target proteins were obtained. The nine key compounds were linolenic acid (1); ethyl-α-d-glucopyranoside (2); 3,5-dihydroxy-6-(hydroxymethyl)oxan-2-one (3); palmitic acid (4); 4-dehydroxy-N-(4,5-methylenedioxy-2-nitrobenzylidene)tyramine (5); pentanoic acid, 3-[(adamantan-1-ylmethyl)carbamoyl]-4-phenyl- (6); phytone (7); 4-Cyclohexene-1,2-dicarboximide, N-butyl-cis- (8); cis,cis,cis-7,10,13-Hexadecatrienal (9). The four target proteins were IL6, MAPK1, P2RX7, and CASP1. MDT was performed between the 11 key compounds and four target proteins to verify network pharmacology results; where the value of MDT was compared with positive controls on each target protein. On MDT, the most potent compounds on IL6 were ethyl-α-d-glucopyranoside and 3,5-dihydroxy-6-(hydroxymethyl)oxan-2-one having an aliphatic heteromonocyclic structure. It was reported that a hetero-aliphatic ring is associated with a better appeal to develop a drug due to its high solubility, low lipophilicity (logP < 5), low albumin-binding and cytochrome P450 inhibition [34,35]. Similarly, Orlistat is a representative anti-obesity drug with an aliphatic heteromonocyclic structure [36] which implies that compounds with an aliphatic heteromonocyclic structure might be potential candidates for anti-obesity drug development.
On the MAPK1 target protein, the most potent compound was 4-dehydroxy-N-(4,5-methylenedioxy-2-nitrobenzylidene)tyramine with an aromatic heteropolycyclic structure. Likewise, Cetilistat is a typical drug for anti-obesity with an aromatic heteropolycyclic structure [36]. On the CASP1 target protein, the most potent compound was pentanoic acid, 3-[(adamantan-1-ylmethyl) carbamoyl] -4-phenyl-) with an aromatic homopolycyclic structure. Moreover, Oleoyl-estrone is a standard drug to reduce the body fat, having an aromatic homopolycyclic structure [36]. Based on these similarities, our study suggests that the four compounds in ICLs have a high chance of offering synergistic effects against obesity.
A bubble chart displayed that ICLs compounds on obesity were involved in 42 target proteins. Furthermore, the outputs of the KEGG pathway enrichment analysis of 42 target proteins indicated that 22 signaling pathways were connected to the progression of obesity, suggesting that these signaling pathways might be the molecular mechanisms of ICLs against obesity. The associations of the 22 signaling pathways with obesity were discussed as follows.
PPAR (peroxisome proliferator-activated receptor) signaling pathway: PPARs are ligand-regulated receptors, and many standard anti-obesity drugs are related to this signaling pathway [37]. VEGF (vascular endothelial growth factor) signaling pathway: VEGF-A (vascular endothelial growth factor A) has anti-inflammatory effects against diet-induced obesity [38]. HIF-1 (hypoxia-inducible factor 1) signaling pathway: inactivation of HIF-1 in adipose tissue alleviates obesity, suggesting that HIF-1 is a new target to develop anti-obesity agents [39]. Fc epsilon RI signaling pathway: an animal experiment demonstrated that mice with obesity increased the expression level of Fc epsilon RI more than eight times as compared to lean mice [40]. It implies that the inactivation of Fc epsilon RI can inhibit obesity. Prolactin signaling pathway: a report shows that prolactin accelerates fat accumulation in diverse animal models; particularly, an increased level of PRL was recorded for obese women in accordance with the visceral fat amount [41]. Interleukin 17 (IL17) signaling pathway: IL17 expression level is enhanced in obese individuals, a mediator to induce pro-inflammatory reactions [42,43]. AGE-RAGE signaling pathway in diabetic complications: AGE-RAGE is deeply interconnected to obesity-involved renal damage; both AGE and RAGE induce a pro-inflammatory reaction and are associated with obesity [44,45]. Tumor necrosis factor (TNF) signaling pathway: TNF inhibits lipoprotein lipase to break triglyceride, known as a primary factor of obesity [46]. Calcium signaling pathway: the activation of calcium signaling promotes energy consumption, which facilitates metabolism and differentiation of adipocytes, thus preventing obesity [47]. Thyroid signaling pathway: obesity is evidently related to Hashimoto’s thyroiditis, suggesting that prevention of obesity is significant for recovering thyroid function [48]. B cell receptor signaling pathway: B cells aggravate obesity-related metabolic disorders and secrete cytokines to stimulate inflammation [49]. T cell receptor signaling pathway: T cell damage is accelerated by obesity; accordingly, T cell dysfunction is detrimental to maintain the immune system [50,51]. Relaxin signaling pathway: the activation of Relaxin-2 attenuates obesity in high-fat-diet mice; furthermore, Relaxin-3 associated with hypercholesterolemia is a potential target protein against obesity [52,53]. Insulin signaling pathway: insulin resistance is a crucial factor in aggravating obesity; especially, adipose tissues in obese individuals produce pro-inflammatory agents that stimulate progressive insulin resistance [54]. Rap-1 signaling pathway: an animal experiment demonstrated that the lack of Rap-1 induces weight gain due to abdominal fat accumulation [55]. Gonadotropin-releasing hormone (GnRH) signaling pathway: GnRH agonist treatment induces fat accumulation; mainly, inhibition of GnRH is a preventive method to treat obesity [56]. Renin-angiotensin system (RAS) signaling pathway: obesity is linked to RAS activation; in contrast, blockers of RAS diminished the type 2 diabetes by 22% in severe populations [57]. Oxytocin signaling pathway: the insufficiency of oxytocin and/or its receptor expression leads to obesity, which is implicated in metabolic disorders [58]. Phosphoinositide 3-Kinase—Protein kinase B (PI3K-AKT) signaling pathway: the dysfunction of the PI3K-AKT signaling pathway causes obesity, in other words, inhibition of the PI3K-AKT signaling pathway exacerbates metabolic processes [59]. Janus kinase/signal transducer and activator of transcription proteins (JAK-STAT) signaling pathway: the JAK-STAT signaling pathway is involved in preventing metabolic diseases including obesity, suggesting that the JAK-STAT pathway is a potential therapeutic mechanism for the treatment of obesity [60]. MAPK signaling pathway: a study indicated that obese mice have shown activated MAPK (known as ERK) expression, while the blocking of MAPK diminishes lipolysis in both mice and human adipose tissue [61]. NOD-like receptor (NLR) signaling pathway: the NLR pathway is overexpressed in the adipocytes from the obesity which increases inflammasome activity [62]. On NOD-like signaling pathway in KEGG pathway enrichment, each CASP1, MAPK1, and IL6 target protein is associated with proinflammatory reactions. In detail, CASP1, MAPK1, and IL6 are deeply involved in metabolic diseases, and their activation leads to obesity-related diseases [63].
According to the degree value of each target protein in the PPI network, IL6 was regarded as a key target of ICLs against obesity, which was directly connected to 52 out of 118 target proteins. In addition, based on the degree value of each target protein in the STC network, MAPK1 was considered as an uppermost target of ICLs against obesity, which was enriched in 20 out of 22 signaling pathways. Specifically, both the MAPK signaling pathway and the NLR signaling pathway had the same rich factor of 0.024. Between the two signaling pathways, a signaling pathway associated with IL6 and MAPK1 was the NLR signaling pathway. Thus, the NLR signaling pathway might be the key signaling pathway of ICLs against obesity. The four target proteins associated with the NLR signaling pathway were IL6, MAPK1, P2RX7, and MAPK1. The four target proteins were used to perform MDT with ligands connected to each target protein; also, MDT was conducted for positive controls to compare each affinity with ligands from ICLs. From the MDT, P2RX7 was excluded due to invalid binding energy (< −6.0 kcal/mol). Each ligand from ICLs bound to three other target proteins (IL6, MAPK1, and CASP1) and exposed higher affinity than the positive controls. Thus, these results suggest that inhibition of the three targets on the NLR signaling pathway might develop a synergistic effect to alleviate the obesity. Our research shows that four compounds, including ethyl- α-d-glucopyranoside (1); 3,5-dihydroxy-6-(hydroxymethyl)oxan-2-one (2); 4-dehydroxy-N-(4,5-methylenedioxy-2-nitrobenzylidenetyramine (3); and pentanoic acid, 3-[(adamantan-1-ylmethyl) carbamoyl] -4-phenyl- (4) from ICLs were noted as promising ligands on the three targets (IL6, MAPK1,and CASP1) (Figure 9).
Figure 9.
Summary figure of key findings in the study.
4. Materials and Methods
4.1. Plant Material Collection and Classification
The Ilex cornuta leaves (ICLs) were collected from Mihogil of Bomunmyeon (Latitude: 35.4023, Longitude: 126.3215), Jeollabuk-do, Republic of Korea, in October 2020, and the plant was identified by Dr. Dong Ha Cho, Plant Biologist and Professor at the Department of Bio-Health Convergence, College of Biomedical Science, Kangwon National University. A voucher number (KNL 011) was deposited at the Kenaf Corporation in the Department of Bio-Health Convergence, and the material can only be used for research purposes.
4.2. Plant Preparation and Extraction
The ICLs were dried in a shady area at room temperature (20–22 °C) for 7 days, and dried leaves were powdered using an electric blender. Approximately 30 g of C. maackii flower powder was soaked in 500 mL of 100% methanol (Daejung, Korea) for 7 days and repeated 3 times for the highest extraction. The solvent extract was collected, filtered, and evaporated using a vacuum evaporator (IKA- RV8, Japan). The evaporated sample was dried under a boiling water bath (IKA-HB10, Japan) at 40 °C to obtain the yield.
4.3. GC-MS Analysis Condition
Agilent 7890A was used to carry out the GC-MS analysis. The GC was equipped with a DB-5 (30 m × 0.25 mm × 0.25 μm) capillary column. Initially, the instrument was maintained at a temperature of 100 °C for 2.1 min. The temperature was increased to 300 °C at the rate of 25 °C/min and maintained for 20 min. The injection port temperature and helium flow rate were confirmed as 250 °C and 1.5 mL/min, respectively. The ionization voltage was 70 eV. The samples were injected in split mode at 10:1. The MS scan range was set at 35–900 (m/z). The fragmentation patterns of mass spectra were compared with those stored in the W8N05ST Library MS database. The percentage of each compound was calculated from the relative peak area of each compound in the chromatogram. The concept of integration used was the ChemStation integrated algorithms.
4.4. Chemical Compounds Database Construction and Drug-Likeness Identification
The chemical compounds from the ICLs leaves were identified through the GC-MS analysis. Then, the GC-MS detected chemical compounds that were filtered by Lipinski’s rule and TPSA value in SwissADME (http://www.swissadme.ch/) (accessed on 23 April 2021) [64] to confirm the ‘drug-likeness’ physicochemical properties. The PubChem repository (https://pubchem.ncbi.nlm.nih.gov/) (accessed on 23 April 2021) was utilized to select the SMILES (simplified molecular input line entry system) format.
4.5. Acquisition of Target Proteins Related to Selected Chemical Compounds or Obesity
Target proteins connected to the bioactives (using SMILES) were selected through both similarity ensemble approach (SEA) (http://sea.bkslab.org/)(accessed on 2 May 2021) [65] and SwissTargetPrediction (STP) (http://www.swisstargetprediction.ch/) (accessed on 2 May 2021) [20] with the ‘Homo Sapiens’ setting. The obesity-related target proteins on humans were obtained from TTD (http://db.idrblab.net/ttd/) (accessed on 6 May 2021) and OMIM (https://www.omim.org/) (accessed on 6 May 2021). The overlapping target proteins between chemical compounds of ICLs and obesity-related target proteins were illustrated by InteractiVenn (http://www.interactivenn.net/) (accessed on 8 May 2021) [66].
4.6. PPI Networks and Signaling Pathways on a Bubble Chart
On the final overlapping target proteins, STRING (https://string-db.org/) (accessed on 11 May 2021) [67] was utilized to analyze the PPI networks. Thereby, RPackage was used to identify the degree of value, which is defined as the numbers of connectivity to a target protein (node). Then, signaling pathways directly related to obesity were visualized on a bubble chart via RPackage. Thus, the signaling pathways provide important clues for the therapeutic effect of ICLs against obesity.
4.7. A Signaling Pathways-Target Proteins-Chemical Compounds Network
The signaling pathways-target proteins-chemical compounds (STC) network was utilized to construct a size map, based on the degree of values. In the network, green circles (nodes) represented signaling pathways; pink rectangles (nodes) represented target proteins, and orange triangles (nodes) represented chemical compounds. The size of the pink rectangles stood for the number of connectivity with signaling pathways; the size of the orange triangles stood for the number of connectivity with target proteins. The merged networks were constructed using RPackage.
4.8. Preparation of Target Proteins for MDT
The target proteins of two key signaling pathways (MAPK signaling pathway, NOD-like receptor signaling pathway), i.e., FGF1 (PDB ID: 3OJ2), FGF2 (PDB ID: 1IIL), PLA2G4A (PDB ID: 1BCI), VEGFA (PDB ID: 3V2A), PRKCA (PDB ID: 3IW4), CASP3 (PDB ID: 5I9B), IL6 (PDB ID: 4NI9), MAPK1 (PDB ID: 4IZ5), P2RX7 (PDB ID: 5U2H), and CASP1 (PDB ID: 3D6F) were selected on STRING via RCSB PDB (https://www.rcsb.org/) (accessed on 12 May 2021). Thus, the target proteins selected as .pdb format were converted into .pdbqt format via Autodock (http://autodock.scripps.edu/) (accessed on 12 May 2021).
4.9. Preparation of Ligands for MDT
The ligand molecules were converted to .sdf from PubChem into .pdb format using Pymol, and the ligand molecules were converted into .pdbqt format through Autodock.
4.10. Preparation of Positive Standard Ligands on the NLR Signaling Pathway for MDT
The number of two positive standard ligands on IL6 (PDB ID: 4NI9) antagonists, i.e., veratric acid (PubChem ID: 7121); the number of one positive ligand on MAPK1 antagonist (PDB ID: 4IZ5), i.e., CU-Cpt 22 (PubChem ID: 71503400); the number of one positive ligand on P2RX7 (PDB ID: 5U2H), i.e., KN-62 (PubChem ID: 5312126); the number of three positive ligands on CASP1 (PDB ID: 3D6F), i.e., belnacasan (PubChem ID: 11398092), mulberroside A (PubChem ID: 6443484), and Q-VD-Oph (PubChem ID: 24794416) were selected to perform MDT.
4.11. Preparation of Ligand Molecules for MDT
The ligand molecules were converted to .sdf from PubChem into .pdb format using Pymol, and the ligand molecules were converted into .pdbqt format through Autodock.
4.12. Ligand-Protein Docking
The ligand molecules were docked with target proteins utilizing autodock4 by setting-up 4 energy range and 8 exhaustiveness as default to obtain 10 different poses of ligand molecules [68]. The active site’s grid box size was x = 40 Å, y = 40Å, and z = 40Å. The 2D binding interactions were used with LigPlot+ v.2.2 (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/) (accessed on 14 May 2021) [69]. After docking, ligands of the lowest binding energy (highest affinity) were selected to visualize the ligand-protein interaction in Pymol.
4.13. Toxicological Properties Prediction by admetSAR
Toxicological properties of key ligands from ICLs were demonstrated utilizing the admetSAR web-service tool (http://lmmd.ecust.edu.cn/admetsar1/predict/) (accessed on 14 May 2021) [70] because toxicity is a critical factor in developing new drugs. Hence, Ames toxicity, carcinogenic properties, acute oral toxicity, and rat acute toxicity were predicted by admetSAR.
5. Conclusions
In conclusion, we firstly analyzed the ‘multi-signaling pathways—multi-target proteins—multi-compounds’ network of ICLs against obesity via MDT. As a result, we found a key signaling pathway (NLR signaling pathway), three target proteins (IL6, MAPK1, and CASP1), and four compounds (ethyl-α-d-glucopyranoside; 3, 5-dihydroxy-6-(hydroxymethyl)oxan-2-one; 4-dehydroxy-N-(4,5-methylenedioxy-2 nitrobenzylidene) tyramine; and pentanoic acid, 3-[(adamantan-1-ylmethyl)carbamoyl]-4-phenyl-). Furthermore, the potential four compounds from ICLs have better affinity on each target protein than positive controls, suggesting that the compounds might be a new agent against obesity. Therefore, our research approach would be valuable for facilitating studies of herbal plants against obesity through network pharmacology.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pr9071106/s1. Table S1: the list of 525, 576, and 219 targets from SEA, STP, and overlapping target proteins between SEA and STP, respectively. Table S2: the list of 3,028 target proteins related to obesity. Table S3: the number of final 118 target proteins of ICLs on obesity.
Author Contributions
Conceptualization, methodology, formal analysis, investigation, data Curation, writing—original draft, K.-K.O.; software, investigation, data curation, K.-K.O. and M.A.; validation, writing—review & editing, M.A.; supervision, project administration, D.-H.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Acknowledgments
This research was acknowledged by the Department of Bio-Health Convergence, College of Biomedical Science, Kangwon National University, Chuncheon 24341, Republic of Korea.
Conflicts of Interest
There is no conflict of interest declared.
Abbreviation
| GnRH | Gonadotropin- Releasing Hormone |
| ICLs | Ilex cornuta leaves |
| IL17 | Interleukin 17 |
| Janus Kinase | Signal Transducer and Activator of Transcription proteins: JAK-STAT |
| MAPK1 | Mitogen-activated protein kinase 1 |
| MDT | Molecular Docking Test |
| NLR | NOD-like receptor |
| OTPs | Overlapping Target Proteins |
| ORTPs | Obesity-Related Target Proteins |
| PhosphoInositide | 3-Kinase—Protein kinase B: PI3K-AKT |
| PPI | Protein–Protein Interaction |
| RAP1 | Ras-proximate-1 |
| SEA | Similarity Ensemble Approach |
| SMILES | Simplified Molecular Input Line Entry System |
| STC | Signaling pathways -Target proteins-Compounds |
| STP | SwissTargetPrediction |
| TNF | Tumor Necrosis Factor |
References
- Hurt, R.T.; Kulisek, C.; Buchanan, L.A.; McClave, S.A. The obesity epidemic: Challenges, health initiatives, and implications for gastroenterologists. Gastroenterol. Hepatol. 2010, 6, 780–792. [Google Scholar]
- Hryhorczuk, C.; Sharma, S.; Fulton, S.E. Metabolic disturbances connecting obesity and depression. Front. Neurosci. 2013, 7, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, K.K.; Adnan, M.; Ju, I.; Cho, D.H. A network pharmacology study on main chemical compounds fromHibiscus cannabinusL. leaves. RSC Adv. 2021, 11, 11062–11082. [Google Scholar] [CrossRef]
- Artham, S.M.; Lavie, C.J.; Milani, R.V.; Ventura, H.O. Obesity and hypertension, heart failure, and coronary heart disease—Risk factor, paradox, and recommendations for weight loss. Ochsner J. 2009, 9, 124–132. [Google Scholar]
- Shi, J.; Fan, J.; Su, Q.; Yang, Z. Cytokines and Abnormal Glucose and Lipid Metabolism. Front. Endocrinol. 2019, 10, 703. [Google Scholar] [CrossRef]
- Rodgers, R.J.; Tschöp, M.H.; Wilding, J.P.H. Anti-obesity drugs: Past, present and future. DMM Dis. Models Mech. 2012, 5, 621–626. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.G.; Park, C.Y. Anti-obesity drugs: A review about their effects and safety. Diabetes Metab. J. 2012, 36, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Chy, M.N.U.; Adnan, M.; Chowdhury, M.R.; Pagano, E.; Kamal, A.T.M.M.; Oh, K.K.; Cho, D.H.; Capasso, R. Central and peripheral pain intervention by Ophiorrhiza rugosa leaves: Potential underlying mechanisms and insight into the role of pain modulators. J. Ethnopharmacol. 2021, 276, 114182. [Google Scholar]
- Sun, N.N.; Wu, T.Y.; Chau, C.F. Natural dietary and herbal products in anti-obesity treatment. Molecules 2016, 21, 1351. [Google Scholar] [CrossRef]
- KR101876617B1—Composition Comprising Extract of Ilex Cornuta for Promoting the Differentiation to Adipocytic Cells—Google Patents. Available online: https://patents.google.com/patent/KR101876617B1/en (accessed on 26 April 2021).
- Kim, J.; Kang, W.; Min, H. In Vitro Anti-Inflammatory Activity of Ilex cornuta Extract Mediated by Inhibition of Extracellular Signal-Regulated Kinase Phosphorylation. J. Med. Food 2017, 20, 981–988. [Google Scholar] [CrossRef]
- Ahn, E.M.; Asamenew, G.; Kim, H.W.; Lee, S.H.; Yoo, S.M.; Cho, S.M.; Cha, Y.S.; Kang, M.S. Anti-obesity effects of petasites japonicus (Meowi) ethanol extract on raw 264.7 macrophages and 3t3-l1 adipocytes and its characterization of polyphenolic compounds. Nutrients 2020, 12, 1261. [Google Scholar] [CrossRef]
- Hao, D.; Gu, X.; Xiao, P.; Liang, Z.; Xu, L.; Peng, Y. Research progress in the phytochemistry and biology of Ilex pharmaceutical resources. Acta Pharm. Sin. B 2013, 3, 8–19. [Google Scholar] [CrossRef] [Green Version]
- Oh, K.K.; Adnan, M.; Cho, D.H. Network pharmacology approach to decipher signaling pathways associated with target proteins of NSAIDs against COVID-19. Sci. Rep. 2021, 11, 9606. [Google Scholar] [CrossRef]
- Chandran, U.; Mehendale, N.; Patil, S.; Chaguturu, R.; Patwardhan, B. Network Pharmacology. In Innovative Approaches in Drug Discovery: Ethnopharmacology, Systems Biology and Holistic Targeting; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 127–164. ISBN 9780128018224. [Google Scholar]
- Oh, K.K.; Adnan, M.; Cho, D.H. Network pharmacology of bioactives from Sorghum bicolor with targets related to diabetes mellitus. PLoS ONE 2020, 15, e0240873. [Google Scholar] [CrossRef]
- Oh, K.K.; Adnan, M.; Cho, D.H. Active ingredients and mechanisms of Phellinus linteus (grown on Rosa multiflora) for alleviation of Type 2 diabetes mellitus through network pharmacology. Gene 2020, 768, 145320. [Google Scholar] [CrossRef]
- Shen, L.; Chen, W.; Zhang, B.; Liu, L.; Cao, Y. Integrating network pharmacology and bioinformatics analysis to explore the mechanism of Yupingfengsan in treating lung adenocarcinoma. Eur. J. Integr. Med. 2019, 31, 100967. [Google Scholar] [CrossRef]
- Wang, Z.; Liang, L.; Yin, Z.; Lin, J. Improving chemical similarity ensemble approach in target prediction. J. Cheminformatics 2016, 8, 20. [Google Scholar] [CrossRef] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W3664. [Google Scholar] [CrossRef] [Green Version]
- Piñero, J.; Queralt-Rosinach, N.; Bravo, À.; Deu-Pons, J.; Bauer-Mehren, A.; Baron, M.; Sanz, F.; Furlong, L.I. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database 2015, 2015, 1–17. [Google Scholar] [CrossRef]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005, 33, D514. [Google Scholar] [CrossRef]
- Zeng, X.; Zhang, P.; He, W.; Qin, C.; Chen, S.; Tao, L.; Wang, Y.; Tan, Y.; Gao, D.; Wang, B.; et al. NPASS: Natural product activity and species source database for natural product research, discovery and tool development. Nucleic Acids Res. 2018, 46, D1217–D1222. [Google Scholar] [CrossRef] [Green Version]
- Hilgren, J.D.; Salverda, J.A. Antimicrobial Efficacy of a Peroxyacetic/Octanoic Acid Mixture in Fresh-Cut-Vegetable Process Waters. J. Food Sci. 2000, 65, 1376–1379. [Google Scholar] [CrossRef]
- Saleem, M. Lupeol, a novel anti-inflammatory and anti-cancer dietary triterpene. Cancer Lett. 2009, 285, 109–115. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Sun, H.; Liu, Y.; Shiroshita, T.; Kawano, S.; Takeshi, S.; Ma, J.; Zhang, Z. Anti-cancer activity comparisons of aqueous extracts from Inonotus obliquus, Cordyceps militaris and Uncaria tomentosa in vitro and in vivo. J. Pharmacogn. Phytochem. 2014, 2, 19–25. [Google Scholar]
- Wang, Q.B.; Sun, L.Y.; Du, Y. Veratric Acid Inhibits LPS-Induced IL-6 and IL-8 Production in Human Gingival Fibroblasts. Inflammation 2016, 39, 237–242. [Google Scholar] [CrossRef]
- Tamai, R.; Suzuki, K.; Mashima, I.; Kiyoura, Y. MPMBP down-regulates Toll-like receptor (TLR) 2 ligand-induced proinflammatory cytokine production by inhibiting NF-κB but not AP-1 activation. Int. Immunopharmacol. 2020, 79. [Google Scholar] [CrossRef]
- Shityakov, S.; Förster, C. In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Adv. Appl. Bioinform. Chem. 2014. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Yang, Q.; He, Y.; Zou, X.; Cao, Z. BmK NT1-induced neurotoxicity is mediated by PKC/CaMKⅡ-dependent ERK1/2 and p38 activation in primary cultured cerebellar granule cells. Toxicology 2019, 421, 22–29. [Google Scholar] [CrossRef]
- Xu, S.; Li, X.; Liu, Y.; Xia, Y.; Chang, R.; Zhang, C. Inflammasome inhibitors: Promising therapeutic approaches against cancer. J. Hematol. Oncol. 2019, 12, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.-C.; Ho, H.-H.; Lin, M.-C.; Huang, C.-N.; Huang, H.-P.; Wang, C.-J. Impact of polyphenolic components from mulberry on apoptosis of vascular smooth muscle cells. J. Sci. Food Agric. 2016, 96, 381–391. [Google Scholar] [CrossRef]
- Renolleau, S.; Fau, S.; Goyenvalle, C.; Joly, L.M.; Chauvier, D.; Jacotot, E.; Mariani, J.; Charriaut-Marlangue, C. Specific caspase inhibitor Q-VD-OPh prevents neonatal stroke in P7 rat: A role for gender. J. Neurochem. 2007, 100, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Aldeghi, M.; Malhotra, S.; Selwood, D.L.; Chan, A.W.E. Two- and three-dimensional rings in drugs. Chem. Biol. Drug Des. 2014, 83, 450–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Li, H.; Tian, L.; Li, Q.; Luo, J.; Zhang, Y. Analysis of the Physicochemical Properties of Acaricides Based on Lipinski’s Rule of Five. J. Comput. Biol. 2020, 27, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34. [Google Scholar] [CrossRef]
- Blaschke, F.; Takata, Y.; Caglayan, E.; Law, R.E.; Hsueh, W.A. Obesity, peroxisome proliferator-activated receptor, and atherosclerosis in type 2 diabetes. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 28–40. [Google Scholar] [CrossRef] [Green Version]
- Elias, I.; Franckhauser, S.; Bosch, F. New insights into adipose tissue VEGF-A actions in the control of obesity and insulin resistance. Adipocyte 2013, 2, 109–112. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Qu, A.; Matsubara, T.; Chanturiya, T.; Jou, W.; Gavrilova, O.; Shah, Y.M.; Gonzalez, F.J. Disruption of hypoxia-inducible factor 1 in adipocytes improves insulin sensitivity and decreases adiposity in high-fat diet-fed mice. Diabetes 2011, 60, 2484–2495. [Google Scholar] [CrossRef] [Green Version]
- Nadler, S.T.; Stoehr, J.P.; Schueler, K.L.; Tanimoto, G.; Yandell, B.S.; Attie, A.D. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc. Natl. Acad. Sci. USA 2000, 97, 11371–11376. [Google Scholar] [CrossRef] [Green Version]
- Kok, P.; Roelfsema, F.; Frölich, M.; Meinders, A.E.; Pijl, H. Prolactin Release Is Enhanced in Proportion to Excess Visceral Fat in Obese Women. J. Clin. Endocrinol. Metab. 2004, 89, 4445–4449. [Google Scholar] [CrossRef]
- Ahmed, M.; Gaffen, S.L. IL-17 in obesity and adipogenesis. Cytokine Growth Factor Rev. 2010, 21, 449–453. [Google Scholar] [CrossRef] [Green Version]
- Zúñiga, L.A.; Shen, W.-J.; Joyce-Shaikh, B.; Pyatnova, E.A.; Richards, A.G.; Thom, C.; Andrade, S.M.; Cua, D.J.; Kraemer, F.B.; Butcher, E.C. IL-17 Regulates Adipogenesis, Glucose Homeostasis, and Obesity. J. Immunol. 2010, 185, 6947–6959. [Google Scholar] [CrossRef] [Green Version]
- Tomino, Y.; Hagiwara, S.; Gohda, T. AGE-RAGE interaction and oxidative stress in obesity-related renal dysfunction. Kidney International 2011, 80, 133–135. [Google Scholar] [CrossRef] [Green Version]
- Leuner, B.; Max, M.; Thamm, K.; Kausler, C.; Yakobus, Y.; Bierhaus, A.; Sel, S.; Hofmann, B.; Silber, R.E.; Simm, A.; et al. RAGE beeinflusst adipositas bei mäusen. einfluss des rezeptors für AGEs (RAGE) auf gewichtszunahme, insulin-und AGE-akkumulation in mäusen unter hochfettdiät. Zeitschrift für Gerontologie und Geriatrie 2012, 45, 102–108. [Google Scholar] [CrossRef]
- Kern, P.A.; Ranganathan, S.; Li, C.; Wood, L.; Ranganathan, G. Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am. J. Physiol. Endocrinol. Metab. 2001, 280, 745–751. [Google Scholar] [CrossRef]
- Song, Z.; Wang, Y.; Zhang, F.; Yao, F.; Sun, C. Calcium signaling pathways: Key pathways in the regulation of obesity. Int. J. Mol. Sci. 2019, 20, 2768. [Google Scholar] [CrossRef] [Green Version]
- Song, R.H.; Wang, B.; Yao, Q.M.; Li, Q.; Jia, X.; Zhang, J.A. The Impact of Obesity on Thyroid Autoimmunity and Dysfunction: A Systematic Review and Meta-Analysis. Front. Immunol. 2019, 10, 2349. [Google Scholar] [CrossRef]
- Liu, R.; Nikolajczyk, B.S. Tissue immune cells fuel obesity-associated inflammation in adipose tissue and beyond. Front. Immunol. 2019, 10, 1587. [Google Scholar] [CrossRef] [Green Version]
- Shirakawa, K.; Yan, X.; Shinmura, K.; Endo, J.; Kataoka, M.; Katsumata, Y.; Yamamoto, T.; Anzai, A.; Isobe, S.; Yoshida, N.; et al. Obesity accelerates T cell senescence in murine visceral adipose tissue. J. Clin. Investig. 2016, 126, 4626–4639. [Google Scholar] [CrossRef] [Green Version]
- Aguilar, E.G.; Murphy, W.J. Obesity induced T cell dysfunction and implications for cancer immunotherapy. Curr. Opin. Immunol. 2018, 51, 181–186. [Google Scholar] [CrossRef]
- Lee, K.C.; Hsieh, Y.C.; Chan, C.C.; Sun, H.J.; Huang, Y.H.; Hou, M.C.; Lin, H.C. Human relaxin-2 attenuates hepatic steatosis and fibrosis in mice with non-alcoholic fatty liver disease. Lab. Investig. 2019, 99, 1203–1216. [Google Scholar] [CrossRef]
- Smith, C.M.; Walker, A.W.; Hosken, I.T.; Chua, B.E.; Zhang, C.; Haidar, M.; Gundlach, A.L. Relaxin-3/RXFP3 networks: An emerging target for the treatment of depression and other neuropsychiatric diseases? Front. Pharmacol. 2014, 5, 46. [Google Scholar] [CrossRef] [Green Version]
- Kahn, S.E.; Hull, R.L.; Utzschneider, K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006, 444, 840–846. [Google Scholar] [CrossRef]
- Martínez, P.; Gómez-López, G.; García, F.; Mercken, E.; Mitchell, S.; Flores, J.M.; deCabo, R.; Blasco, M.A. RAP1 Protects from Obesity through Its Extratelomeric Role Regulating Gene Expression. Cell Rep. 2013, 3, 2059–2074. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, H.; Douchi, T.; Yamamoto, S.; Toshimichi, O.; Kuwahata, R.; Nagata, Y. Body fat distribution and body composition during GnRH agonist therapy. Obstet. Gynecol. 2001, 97, 338–342. [Google Scholar] [CrossRef]
- Goossens, G.H. The Renin-Angiotensin System in the Pathophysiology of Type 2 Diabetes. Obes. Facts 2012, 5, 611–624. [Google Scholar] [CrossRef]
- Ding, C.; Leow, M.K.S.; Magkos, F. Oxytocin in metabolic homeostasis: Implications for obesity and diabetes management. Obes. Rev. 2019, 20, 22–40. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Liu, G.; Guo, J.; Su, Z.Q. The PI3K/AKT pathway in obesity and type 2 diabetes. Int. J. Biol. Sci. 2018, 14, 1483–1496. [Google Scholar] [CrossRef] [Green Version]
- Dodington, D.W.; Desai, H.R.; Woo, M. JAK/STAT—Emerging Players in Metabolism. Trends Endocrinol. Metab. 2018, 29, 55–65. [Google Scholar] [CrossRef]
- Hong, S.; Song, W.; Zushin, P.J.H.; Liu, B.; Jedrychowski, M.P.; Mina, A.I.; Deng, Z.; Cabarkapa, D.; Hall, J.A.; Palmer, C.J.; et al. Phosphorylation of Beta-3 adrenergic receptor at serine 247 by ERK MAP kinase drives lipolysis in obese adipocytes. Mol. Metab. 2018, 12, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Deng, T.; Peterson, L.E.; Yu, R.; Lin, J.; Hamilton, D.J.; Reardon, P.R.; Sherman, V.; Winnier, G.E.; Zhan, M.; et al. Transcriptome analysis of human adipocytes implicates the NOD-like receptor pathway in obesity-induced adipose inflammation. Mol. Cell. Endocrinol. 2014, 394, 80–87. [Google Scholar] [CrossRef] [Green Version]
- Febbraio, M.A. Role of interleukins in obesity: Implications for metabolic disease. Trends Endocrinol. Metab. 2014, 25, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Khanal, P.; Patil, B.M.; Chand, J.; Naaz, Y. Anthraquinone Derivatives as an Immune Booster and their Therapeutic Option Against COVID-19. Nat. Prod. Bioprospect. 2020, 10, 325–335. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. Ligplot: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).