Ultra-Fast Electrochemical Sensor for Point-of-Care COVID-19 Diagnosis Using Non-Invasive Saliva Sampling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
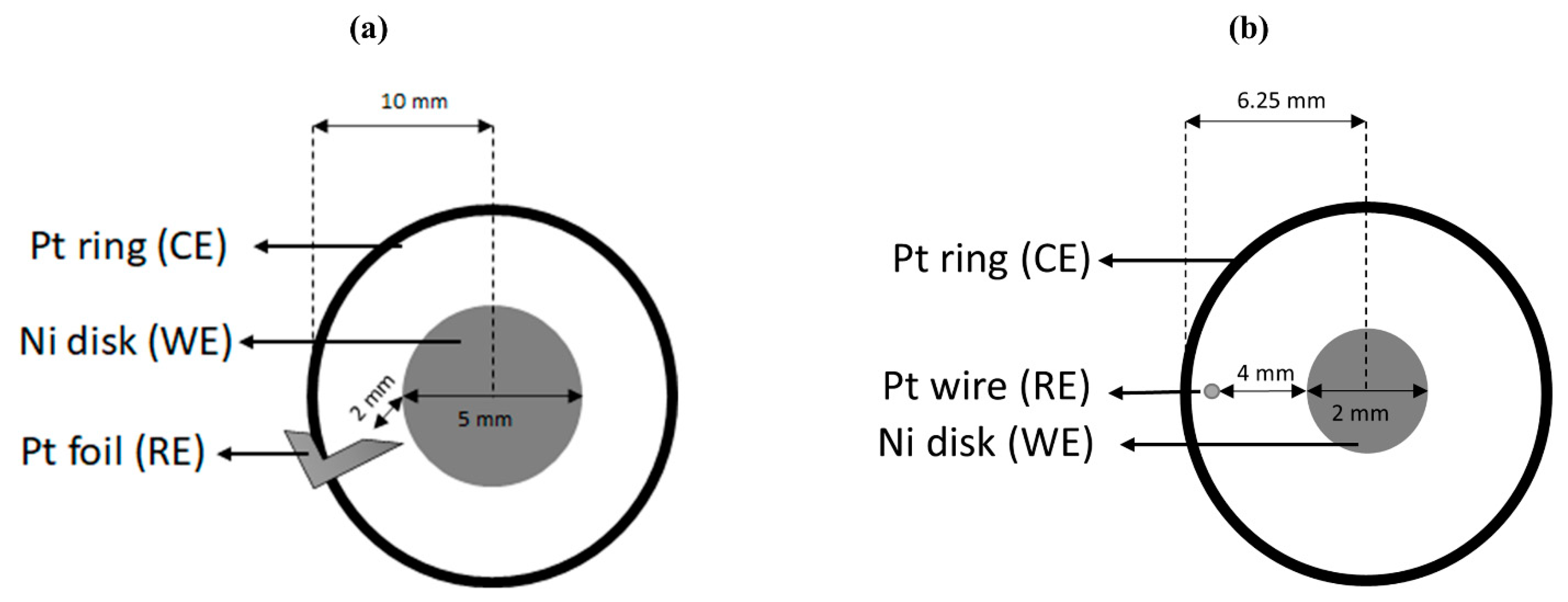
2.2. Electrode Probe Configuration
2.2.1. Rotating Disk Electrode (RDE) Setup
2.2.2. Miniature RDE Setup
2.3. Testing Procedure
2.3.1. Activation
2.3.2. Sample Test
2.3.3. Rinse
3. Results and Discussion
3.1. Calibration Generation
3.2. Calibration Validation
3.3. Specificity
3.4. SARS-CoV-2 Detection in Saliva
3.5. Detection Limit for Practical Use
3.6. Mechanism Hypothesis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, S.; Li, Y. Beware of the Second Wave of COVID-19. Lancet 2020, 395, 1321–1322. [Google Scholar] [CrossRef]
- Kuehn, B.M. Africa Succeeded Against COVID-19’s First Wave, but the Second Wave Brings New Challenges. JAMA 2021, 325, 327. [Google Scholar] [CrossRef] [PubMed]
- Soriano, V.; de Mendoza, C.; Gómez-Gallego, F.; Corral, O.; Barreiro, P. Third Wave of COVID-19 in Madrid, Spain. Int. J. Infect. Dis. 2021, 107, 212–214. [Google Scholar] [CrossRef]
- Taboada, M.; González, M.; Alvarez, A.; Eiras, M.; Costa, J.; Álvarez, J.; Seoane-Pillado, T. First, Second and Third Wave of COVID-19. What Have We Changed in the ICU Management of These Patients? J. Infect. 2021, 82, e14–e15. [Google Scholar] [CrossRef] [PubMed]
- D’Arienzo, M.; Coniglio, A. Assessment of the SARS-CoV-2 Basic Reproduction Number, R0, Based on the Early Phase of COVID-19 Outbreak in Italy. Biosaf. Health 2020. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial Undocumented Infection Facilitates the Rapid Dissemination of Novel Coronavirus (SARS-CoV-2). Science 2020, 368, 489–493. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.I.; Burbelo, P.D. Reinfection with SARS-CoV-2: Implications for Vaccines. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bauza, V.; Sclar, G.D.; Bisoyi, A.; Owens, A.; Ghugey, A.; Clasen, T. Experience of the COVID-19 Pandemic in Rural Odisha, India: Knowledge, Preventative Actions, and Impacts on Daily Life. Int. J. Environ. Res. Public Health 2021, 18, 2863. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061. [Google Scholar] [CrossRef]
- Tang, Y.-W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, e00512-20. [Google Scholar] [CrossRef] [Green Version]
- Dheda, K.; Davids, M.; Chang, J.-W.; Gina, P.; Pooran, A.; Makambwa, E.; Esmail, A.; Vardas, E.; Preiser, W. London School of Hygiene and Tropical Medicine Diagnosis of COVID-19: Considerations, Controversies and Challenges in South Africa. Wits J. Clin. Med. 2020, 2, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Rothman, R.E. PCR-Based Diagnostics for Infectious Diseases: Uses, Limitations, and Future Applications in Acute-Care Settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef]
- Gutierres, S.L.; Welty, T.E. Point-of-Care Testing: An Introduction. Ann. Pharmacother. 2004, 38, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhong, Z.; Zhao, W.; Zheng, C.; Wang, F.; Liu, J. Chest CT for Typical Coronavirus Disease 2019 (COVID-19) Pneumonia: Relationship to Negative RT-PCR Testing. Radiology 2020, 296, E41–E45. [Google Scholar] [CrossRef] [Green Version]
- Shabani, E.; Dowlatshahi, S.; Abdekhodaie, M.J. Laboratory Detection Methods for the Human Coronaviruses. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 225–246. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Ahmed, J.A.; Eidex, R.B.; Nyoka, R.; Waiboci, L.W.; Erdman, D.; Tepo, A.; Mahamud, A.S.; Kabura, W.; Nguhi, M.; et al. Comparison of Nasopharyngeal and Oropharyngeal Swabs for the Diagnosis of Eight Respiratory Viruses by Real-Time Reverse Transcription-PCR Assays. PLoS ONE 2011, 6, e21610. [Google Scholar] [CrossRef] [Green Version]
- Blaschke, A.J.; Allison, M.A.; Meyers, L.; Rogatcheva, M.; Heyrend, C.; Mallin, B.; Carter, M.; LaFleur, B.; Barney, T.; Poritz, M.A.; et al. Non-Invasive Sample Collection for Respiratory Virus Testing by Multiplex PCR. J. Clin. Virol. 2011, 52, 210–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilder, L.; Machtei, E.E.; Shenhar, Y.; Kra-Oz, Z.; Basis, F. Salivary Detection of H1N1 Virus: A Clinical Feasibility Investigation. J. Dent. Res. 2011, 90, 1136–1139. [Google Scholar] [CrossRef] [PubMed]
- Keaney, D.; Whelan, S.; Finn, K.; Lucey, B. Misdiagnosis of SARS-CoV-2: A Critical Review of the Influence of Sampling and Clinical Detection Methods. Med. Sci. 2021, 9, 36. [Google Scholar] [CrossRef]
- Pasomsub, E.; Watcharananan, S.P.; Boonyawat, K.; Janchompoo, P.; Wongtabtim, G.; Suksuwan, W.; Sungkanuparph, S.; Phuphuakrat, A. Saliva Sample as a Non-Invasive Specimen for the Diagnosis of Coronavirus Disease 2019: A Cross-Sectional Study. Clin. Microbiol. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sindhu, S.; Jagannathan, N. Saliva: A Cutting Edge in Diagnostic Procedures. J. Oral Dis. 2014, 2014, 168584. [Google Scholar] [CrossRef] [Green Version]
- Azzi, L.; Carcano, G.; Gianfagna, F.; Grossi, P.; Gasperina, D.D.; Genoni, A.; Fasano, M.; Sessa, F.; Tettamanti, L.; Carinci, F.; et al. Saliva Is a Reliable Tool to Detect SARS-CoV-2. J. Infect. 2020, 81, e45–e50. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.-W.; Tsang, O.T.-Y.; Yip, C.C.-Y.; Chan, K.-H.; Wu, T.-C.; Chan, J.M.-C.; Leung, W.-S.; Chik, T.S.-H.; Choi, C.Y.-C.; Kandamby, D.H.; et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [Green Version]
- Williams, E.; Bond, K.; Zhang, B.; Putland, M.; Williamson, D.A. Saliva as a Non-Invasive Specimen for Detection of SARS-CoV-2. J. Clin. Microbiol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Botte, G.G.; Ramanujam, A. Rapid Viral Diagnostic Sensor. U.S. Patent No. 11,060,995, 13 July 2021. [Google Scholar]
- Ramanujam, A.; Neyhouse, B.; Keogh, R.A.; Muthuvel, M.; Carroll, R.K.; Botte, G.G. Rapid Electrochemical Detection of Escherichia Coli Using Nickel Oxidation Reaction on a Rotating Disk Electrode. Chem. Eng. J. 2021, 411, 128453. [Google Scholar] [CrossRef]
- Vedharathinam, V.; Botte, G.G. Understanding the Electro-Catalytic Oxidation Mechanism of Urea on Nickel Electrodes in Alkaline Medium. Electrochim. Acta 2012, 81, 292–300. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [Green Version]
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Lee, S.Y.; Lertanantawong, B. Rapid Electrochemical Detection of Coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 802. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical Biosensors: Recommended Definitions and Classification *. Anal. Lett. 2001, 34, 635–659. [Google Scholar] [CrossRef] [Green Version]
- Kliger, Y.; Levanon, E.Y. Cloaked Similarity between HIV-1 and SARS-CoV Suggests an Anti-SARS Strategy. BMC Microbiol. 2003, 3, 20. [Google Scholar] [CrossRef] [Green Version]
- Khodamoradi, Z.; Moghadami, M.; Lotfi, M. Co-Infection of Coronavirus Disease 2019 and Influenza A: A Report from Iran. Arch. Iran Med. 2020, 23, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Quinn, J.; Pinsky, B.; Shah, N.H.; Brown, I. Rates of Co-Infection Between SARS-CoV-2 and Other Respiratory Pathogens. JAMA 2020, 323, 2085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konala, V.M.; Adapa, S.; Gayam, V.; Naramala, S.; Daggubati, S.R.; Kammari, C.B.; Chenna, A. Co-Infection with Influenza A and COVID-19. Eur. J. Case Rep. Intern. Med. 2020, 7, 1. [Google Scholar] [CrossRef]
- Altuntas Aydin, O.; Kumbasar Karaosmanoglu, H.; Kart Yasar, K. HIV/SARS-CoV-2 Coinfected Patients in Istanbul, Turkey. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Cao, Y.; Xu, S.; Zhou, M. Co-infection of SARS-CoV-2 and HIV in a Patient in Wuhan City, China. J. Med. Virol. 2020, 92, 529–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, A.; Shaw, J.; Karamchand, S.; Lahri, S.; Schrueder, N.; Chothia, M.-Y.; Mowlana, A.; Lalla, U.; Allwood, B.W.; Koegelenberg, C.F.N.; et al. HIV and SARS-CoV-2 Co-Infection: The Diagnostic Challenges of Dual Pandemics. S. Afr. Med. J. 2020, 110, 473–475. [Google Scholar] [CrossRef]
- Wang, W.-K.; Chen, S.-Y.; Liu, I.-J.; Chen, Y.-C.; Chen, H.-L.; Yang, C.-F.; Chen, P.-J.; Yeh, S.-H.; Kao, C.-L.; Huang, L.-M.; et al. Detection of SARS-Associated Coronavirus in Throat Wash and Saliva in Early Diagnosis. Emerg. Infect. Dis. 2004, 10, 1213–1219. [Google Scholar] [CrossRef]
- Liu, L.; Wei, Q.; Alvarez, X.; Wang, H.; Du, Y.; Zhu, H.; Jiang, H.; Zhou, J.; Lam, P.; Zhang, L.; et al. Epithelial Cells Lining Salivary Gland Ducts Are Early Target Cells of Severe Acute Respiratory Syndrome Coronavirus Infection in the Upper Respiratory Tracts of Rhesus Macaques. J. Virol. 2011, 85, 4025–4030. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Li, Y.; Gan, F.; Du, Y.; Yao, Y. Salivary Glands: Potential Reservoirs for COVID-19 Asymptomatic Infection. J. Dent. Res. 2020, 99, 989. [Google Scholar] [CrossRef] [Green Version]
- Sabino-Silva, R.; Jardim, A.C.G.; Siqueira, W.L. Coronavirus COVID-19 Impacts to Dentistry and Potential Salivary Diagnosis. Clin. Oral Investig. 2020, 24, 1619–1621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bui, V.N.; Nguyen, K.V.; Pham, N.T.; Bui, A.N.; Dao, T.D.; Nguyen, T.T.; Nguyen, H.T.; Trinh, D.Q.; Inui, K.; Uchiumi, H.; et al. Potential of Electrolyzed Water for Disinfection of Foot-and-Mouth Disease Virus. J. Vet. Med. Sci. 2017, 79, 726–729. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, S.P.; Williamson, R.T. A Review of Saliva: Normal Composition, Flow, and Function. J. Prosthet. Dent. 2001, 85, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Park, G.-S.; Moon, J.H.; Ku, K.; Beak, S.-H.; Lee, C.-S.; Kim, S.; Park, E.C.; Park, D.; Lee, J.-H.; et al. Comparative Analysis of Primer–Probe Sets for RT-QPCR of COVID-19 Causative Virus (SARS-CoV-2). ACS Infect. Dis. 2020, 6, 2513–2523. [Google Scholar] [CrossRef]
- Arnaout, R.; Lee, R.A.; Lee, G.R.; Callahan, C.; Yen, C.F.; Smith, K.P.; Arora, R.; Kirby, J.E. SARS-CoV2 Testing: The Limit of Detection Matters. Microbiology 2020. [Google Scholar] [CrossRef]
- Diaz-Morales, O.; Ferrus-Suspedra, D.; Koper, M.T.M. The Importance of Nickel Oxyhydroxide Deprotonation on Its Activity towards Electrochemical Water Oxidation. Chem. Sci. 2016, 7, 2639–2645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Karki, C.B.; Du, D.; Li, H.; Wang, J.; Sobitan, A.; Teng, S.; Tang, Q.; Li, L. Spike Proteins of SARS-CoV and SARS-CoV-2 Utilize Different Mechanisms to Bind With Human ACE2. Front. Mol. Biosci. 2020, 7, 591873. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A.; Botte, G.G. Mathematical Modeling of Ammonia Electrooxidation Kinetics in a Polycrystalline Pt Rotating Disk Electrode. Electrochim. Acta 2015, 179, 519–528. [Google Scholar] [CrossRef] [Green Version]
- Lopin, P.; Lopin, K.V. PSoC-Stat: A Single Chip Open Source Potentiostat Based on a Programmable System on a Chip. PLoS ONE 2018, 13, e0201353. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramanujam, A.; Almodovar, S.; Botte, G.G. Ultra-Fast Electrochemical Sensor for Point-of-Care COVID-19 Diagnosis Using Non-Invasive Saliva Sampling. Processes 2021, 9, 1236. https://doi.org/10.3390/pr9071236
Ramanujam A, Almodovar S, Botte GG. Ultra-Fast Electrochemical Sensor for Point-of-Care COVID-19 Diagnosis Using Non-Invasive Saliva Sampling. Processes. 2021; 9(7):1236. https://doi.org/10.3390/pr9071236
Chicago/Turabian StyleRamanujam, Ashwin, Sharilyn Almodovar, and Gerardine G. Botte. 2021. "Ultra-Fast Electrochemical Sensor for Point-of-Care COVID-19 Diagnosis Using Non-Invasive Saliva Sampling" Processes 9, no. 7: 1236. https://doi.org/10.3390/pr9071236
APA StyleRamanujam, A., Almodovar, S., & Botte, G. G. (2021). Ultra-Fast Electrochemical Sensor for Point-of-Care COVID-19 Diagnosis Using Non-Invasive Saliva Sampling. Processes, 9(7), 1236. https://doi.org/10.3390/pr9071236






