A Denoise Network for Structured Illumination Microscopy with Low-Light Exposure
Abstract
1. Introduction
2. Methods
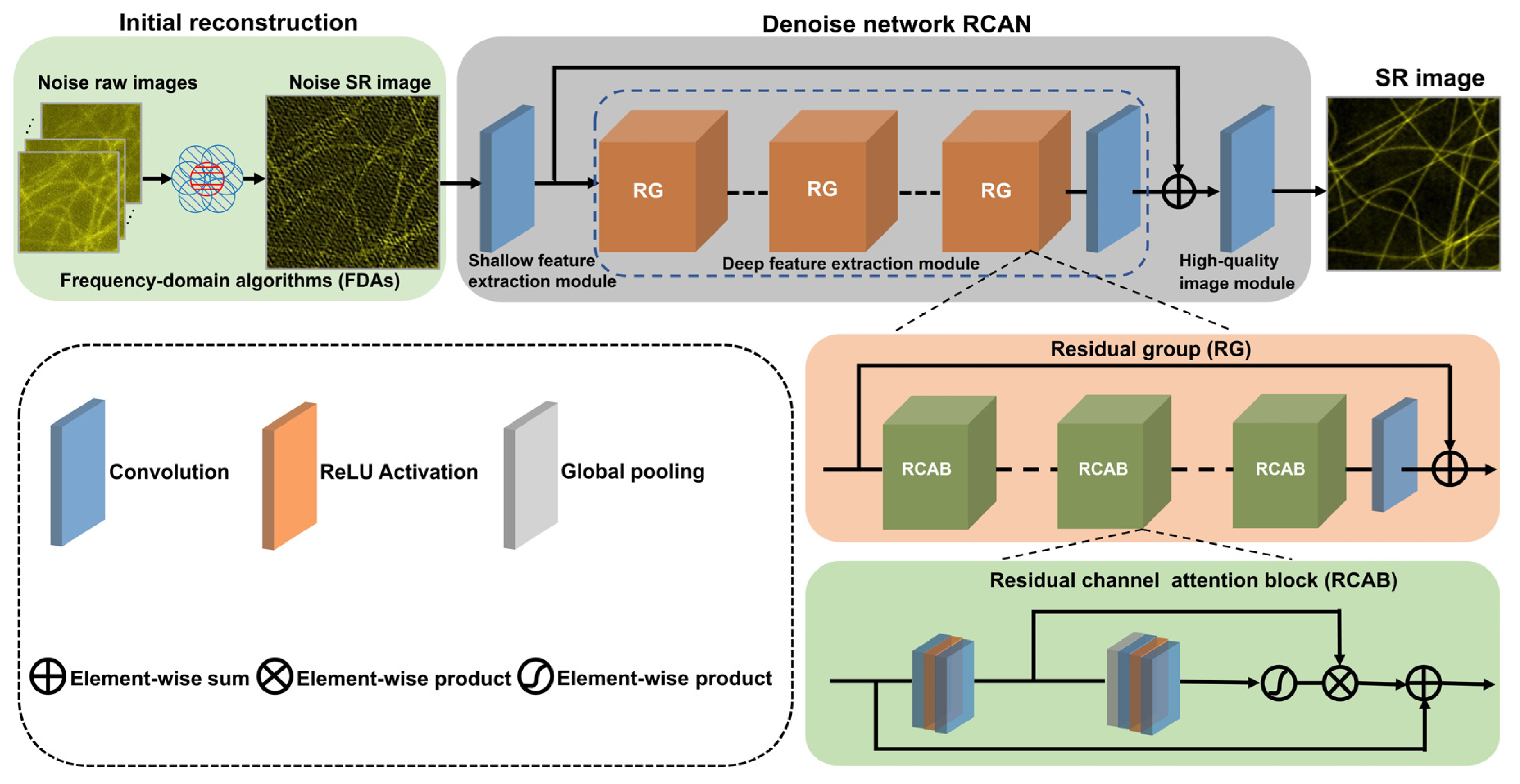
2.1. ND-SIM Network
2.2. Image Assessment Metrics Calculation
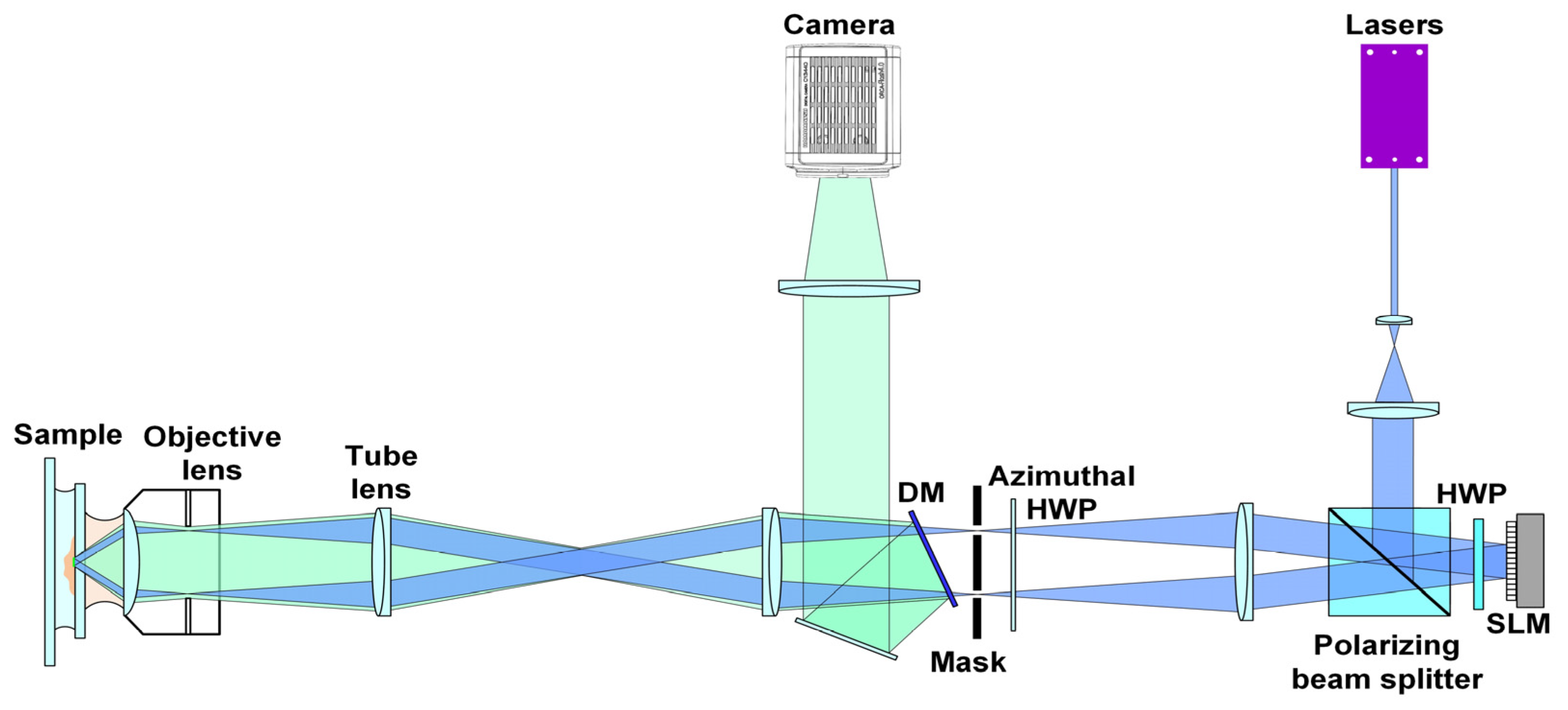
2.3. Data Acquisition of ND-SIM
3. Experimental Results
3.1. Evaluation of ND-SIM in Low-Photon-Count Level
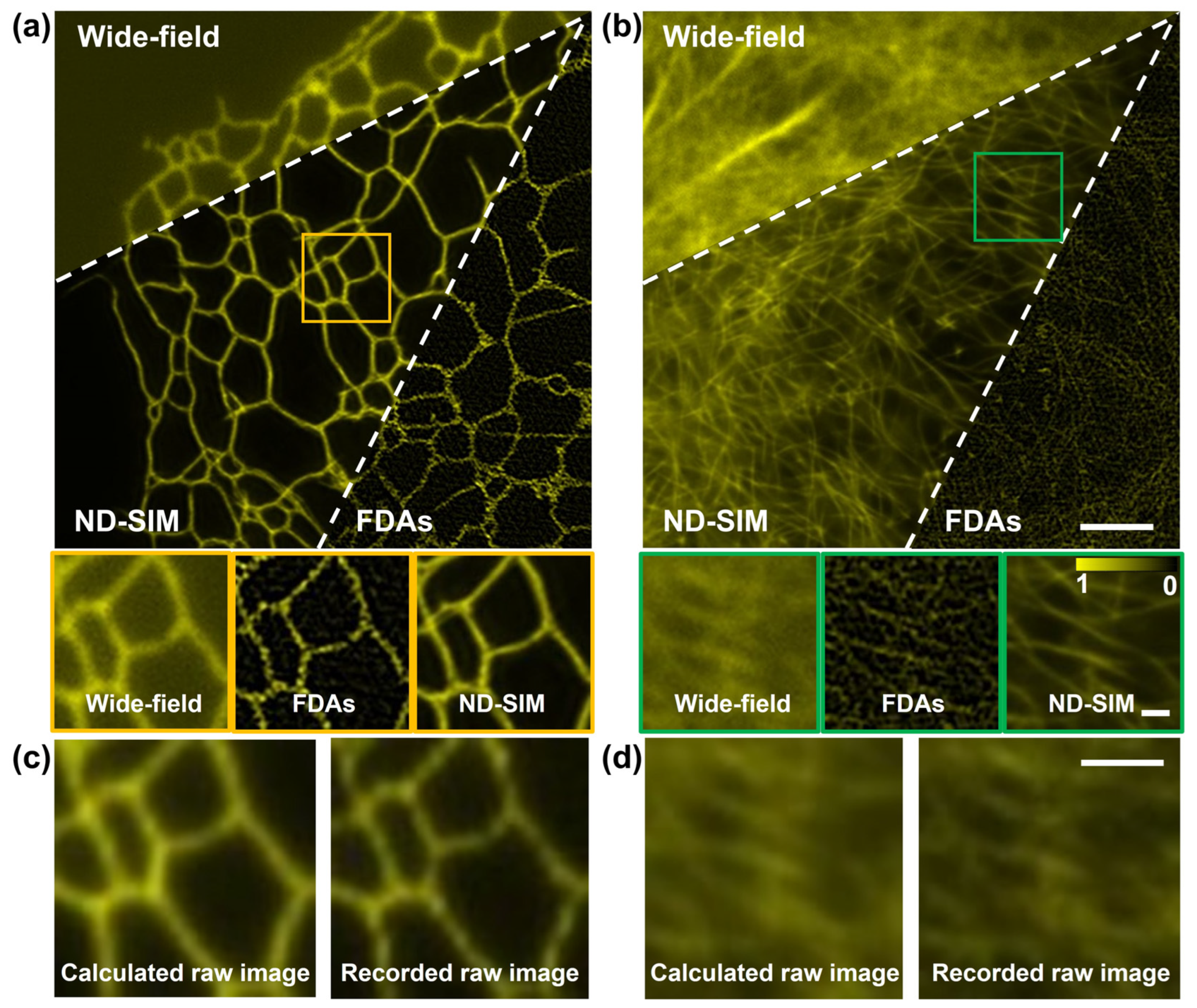
3.2. Comparison of ND-SIM with Other Existing SR Image-Denoising Methods
3.3. ND-SIM Reconstruction for Different Subcellular Organelles
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moore, A.S.; Coscia, S.M.; Simpson, C.L.; Ortega, F.E.; Wait, E.C.; Heddleston, J.M.; Nirschl, J.J.; Obara, C.J.; Guedes-Dias, P.; Boecker, C.A. Actin cables and comet tails organize mitochondrial networks in mitosis. Nature 2021, 591, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.J.; Voeltz, G.K. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 2016, 17, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Prinz, W.A.; Toulmay, A.; Balla, T. The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 2020, 21, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Heintzmann, R.; Cremer, C.G. Laterally modulated excitation microscopy: Improvement of resolution by using a diffraction grating. In Proceedings of the Optical Biopsies and Microscopic Techniques III; Society of Photo Optical: Bellingham, WC, USA, 1999; pp. 185–196. [Google Scholar]
- Blom, H.; Widengren, J. Stimulated emission depletion microscopy. Chem. Rev. 2017, 117, 7377–7427. [Google Scholar] [CrossRef] [PubMed]
- Rego, E.H.; Shao, L.; Macklin, J.J.; Winoto, L.; Johansson, G.A.; Kamps-Hughes, N.; Davidson, M.W.; Gustafsson, M.G. Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proc. Natl. Acad. Sci. USA 2012, 109, E135–E143. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Prunsche, B.; Zhou, L.; Nienhaus, K.; Nienhaus, G.U. Background suppression in fluorescence nanoscopy with stimulated emission double depletion. Nat. Photonics 2017, 11, 163–169. [Google Scholar] [CrossRef]
- Heintzmann, R.; Huser, T. Super-resolution structured illumination microscopy. Chem. Rev. 2017, 117, 13890–13908. [Google Scholar] [CrossRef] [PubMed]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Klar, T.A.; Jakobs, S.; Dyba, M.; Egner, A.; Hell, S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. USA 2000, 97, 8206–8210. [Google Scholar] [CrossRef]
- Huang, X.; Fan, J.; Li, L.; Liu, H.; Wu, R.; Wu, Y.; Wei, L.; Mao, H.; Lal, A.; Xi, P. Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat. Biotechnol. 2018, 36, 451–459. [Google Scholar] [CrossRef]
- Markwirth, A.; Lachetta, M.; Mönkemöller, V.; Heintzmann, R.; Hübner, W.; Huser, T.; Müller, M. Video-rate multi-color structured illumination microscopy with simultaneous real-time reconstruction. Nat. Commun. 2019, 10, 4315. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhong, S.; Hou, Y.; Cao, R.; Wang, W.; Li, D.; Dai, Q.; Kim, D.; Xi, P. Superresolution structured illumination microscopy reconstruction algorithms: A review. Light Sci. Appl. 2023, 12, 172. [Google Scholar] [CrossRef]
- Müller, M.; Mönkemöller, V.; Hennig, S.; Hübner, W.; Huser, T. Open-source image reconstruction of super-resolution structured illumination microscopy data in ImageJ. Nat. Commun. 2016, 7, 10980. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Shan, C.; Xi, P. Structured illumination microscopy image reconstruction algorithm. IEEE J. Sel. Top. Quantum Electron. 2016, 22, 50–63. [Google Scholar] [CrossRef]
- Wen, G.; Li, S.; Wang, L.; Chen, X.; Sun, Z.; Liang, Y.; Jin, X.; Xing, Y.; Jiu, Y.; Tang, Y. High-fidelity structured illumination microscopy by point-spread-function engineering. Light Sci. Appl. 2021, 10, 70. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Cao, Y.; Bi, Y.; Wu, H.; Liu, Y.; Chen, Q.; Zuo, C. Structured illumination microscopy based on principal component analysis. ELight 2023, 3, 4. [Google Scholar] [CrossRef]
- Fan, J.; Huang, X.; Li, L.; Tan, S.; Chen, L. A protocol for structured illumination microscopy with minimal reconstruction artifacts. Biophys. Rep. 2019, 5, 80–90. [Google Scholar] [CrossRef]
- Hoffman, D.P.; Betzig, E. Tiled reconstruction improves structured illumination microscopy. BioRxiv 2020, BioRxiv:2020.01.06.895318. [Google Scholar]
- Zhao, W.; Zhao, S.; Li, L.; Huang, X.; Xing, S.; Zhang, Y.; Qiu, G.; Han, Z.; Shang, Y.; Sun, D.E.; et al. Sparse deconvolution improves the resolution of live-cell super-resolution fluorescence microscopy. Nat. Biotechnol. 2022, 40, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Liu, B.; Zhao, F.; Hahn, S.; Dong, B.; Song, R.; Elston, T.C.; Xu, Y.; Hahn, K.M. Deep learning enables structured illumination microscopy with low light levels and enhanced speed. Nat. Commun. 2020, 11, 1934. [Google Scholar] [CrossRef] [PubMed]
- Christensen, C.N.; Ward, E.N.; Lu, M.; Lio, P.; Kaminski, C.F. ML-SIM: Universal reconstruction of structured illumination microscopy images using transfer learning. Biomed. Opt. Express 2021, 12, 2720–2733. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.; Li, D.; Liu, Y.; Zhang, S.; Liu, K.; Liu, C.; Guo, Y.; Jiang, T.; Fang, C.; Li, N. Rationalized deep learning super-resolution microscopy for sustained live imaging of rapid subcellular processes. Nat. Biotechnol. 2023, 41, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.; Li, D.; Guo, Y.; Liu, C.; Jiang, T.; Dai, Q.; Li, D. Evaluation and development of deep neural networks for image super-resolution in optical microscopy. Nat. Methods 2021, 18, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Rahaman, N.; Baratin, A.; Arpit, D.; Draxler, F.; Lin, M.; Hamprecht, F.; Bengio, Y.; Courville, A. On the spectral bias of neural networks. In International Conference on Machine Learning; PMLR: London, China, 2019; pp. 5301–5310. [Google Scholar]
- Zhang, Y.; Li, K.; Li, K.; Wang, L.; Zhong, B.; Fu, Y. Image super-resolution using very deep residual channel attention networks. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 286–301. [Google Scholar]
- Liu, X.; Li, J.; Li, J.; Ali, N.; Zhao, T.; An, S.; Zheng, J.; Ma, Y.; Qian, J.; Zuo, C. A cascaded deep network for reconstruction of structured illumination microscopy. Opt. Laser Technol. 2024, 170, 110224. [Google Scholar] [CrossRef]
- Descloux, A.; Grußmayer, K.S.; Radenovic, A. Parameter-free image resolution estimation based on decorrelation analysis. Nat. Methods 2019, 16, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Dabov, K.; Foi, A.; Katkovnik, V.; Egiazarian, K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans. Image Process. 2007, 16, 2080–2095. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Y.; Nichols, E.; Wang, Q.; Zhang, S.; Smith, C.; Howard, S. A poisson-gaussian denoising dataset with real fluorescence microscopy images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 11710–11718. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Li, J.; Song, L.; Zhuo, K.; Wen, K.; An, S.; Ma, Y.; Zheng, J.; Gao, P. A Denoise Network for Structured Illumination Microscopy with Low-Light Exposure. Photonics 2024, 11, 776. https://doi.org/10.3390/photonics11080776
Liu X, Li J, Song L, Zhuo K, Wen K, An S, Ma Y, Zheng J, Gao P. A Denoise Network for Structured Illumination Microscopy with Low-Light Exposure. Photonics. 2024; 11(8):776. https://doi.org/10.3390/photonics11080776
Chicago/Turabian StyleLiu, Xin, Jinze Li, Liangfeng Song, Kequn Zhuo, Kai Wen, Sha An, Ying Ma, Juanjuan Zheng, and Peng Gao. 2024. "A Denoise Network for Structured Illumination Microscopy with Low-Light Exposure" Photonics 11, no. 8: 776. https://doi.org/10.3390/photonics11080776
APA StyleLiu, X., Li, J., Song, L., Zhuo, K., Wen, K., An, S., Ma, Y., Zheng, J., & Gao, P. (2024). A Denoise Network for Structured Illumination Microscopy with Low-Light Exposure. Photonics, 11(8), 776. https://doi.org/10.3390/photonics11080776







