Single Nucleotide Polymorphisms and Dental Fluorosis: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Population, Exposure, Control and Outcome (PECO) Strategy, and Eligibility Criteria
2.3. Information Sources
2.4. Search Strategy
2.5. Selection Process
2.6. Data Collection Process
2.7. Data Items
2.8. Methodological Quality Assessment and Risk of Bias
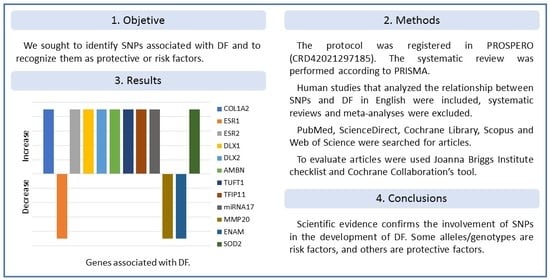
3. Results
3.1. Study Selection
3.2. Study Characteristics
3.3. Methodological Quality Assessment and Risk of Bias
3.4. Results of Individual Studies
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Authors | Gene: SNP | Allele/Genotype | |
|---|---|---|---|
| Huang et al. (2008) [18] | COL1A2: | rs4266 | rr/Rr/RR |
| Ba et al. (2009) [19] | OC: | rs1800247 | hh/Hh/HH |
| Wang et al. (2010) [20] | ESR2: | RsaI | rr/Rr/RR |
| Ba et al. (2011) [21] | ESR1: | rs9340799 | pp/Pp/PP |
| Wen et al. (2012) [22] | PTH: | Bst BI | bb/Bb/BB |
| Escobar-García et al. (2016) [23] | COL1A2: | rs412777 | A/C |
| Küchler et al. (2017) [24] | TIMP2: | rs7501477 | G/T |
| MMP13: | rs2252070 | A/G | |
| Küchler et al. (2018) [25] | ENAM: | rs12640848 | A/G |
| AMELX: | rs946252 | A/G | |
| Dalledone et al. (2019) [27] | ESR1: | rs1884051 | A/G |
| rs9340799 | A/G | ||
| rs2234693 | C/T | ||
| ESR2: | rs4986938 | A/G | |
| rs1256049 | A/G | ||
| ESRRβ: | rs745011 | C/T | |
| rs6574293 | A/G | ||
| rs10132091 | C/T | ||
| rs4903399 | C/T | ||
| rs1077430 | A/G | ||
| rs1676303 | C/T | ||
| Romualdo et al. (2019) [28] | MMP20: | rs1711437 | A/G |
| MMP2: | rs243867 | A/G | |
| Tremillo-Maldonado et al. (2020) [30] | AMELX: | rs946252 | C/T |
| ODAM: | rs1514392 | G | |
| Saha et al. (2021) [33] | ESR1: | rs2234693 | T/C |
| rs2228480 | G/A | ||
| rs3798577 | T/C | ||
| rs2077647 | T/C | ||
| rs9340799 | A/G | ||
| rs2077647 | T/C | ||
| COL1A2: | rs42524 | C/G | |
| rs412777 | A/C | ||
| BGLAP: | rs1800247 | T/C | |
| SPARC: | rs6579885 | G/C | |
| rs4958278 | T/C | ||
| Yuhui et al. (2022) [34] | SOD2: | rs4880 | A/G |
| SOD3: | rs13306703 | C/T | |
| rs2855262 | T/C | ||
| Chakraborty et al. (2022) [35] | ESR1: | rs2077647 | C/T |
| rs2234693 | C/T | ||
| rs9340799 | A/G | ||
| rs2228480 | A/G | ||
| rs3798577 | C/T | ||
| COL1A2: | rs412777 | A/C | |
| rs42524 | C/G | ||
| BGLAP: | rs1543294 | C/T | |
| rs1800247 | C/T | ||
| rs759330 | A/G | ||
| SPARC: | rs4958278 | C/T | |
| rs1800247 | C/G | ||
| rs6579885 | C/T | ||
| VDR: | rs731236 | A/G | |
| MMP20: | rs2285053 | C/T | |
References
- Yadav, K.K.; Kumar, S.; Pham, Q.B.; Gupta, N.; Rezania, S.; Kamyab, H.; Yadav, S.; Vymazal, J.; Kumar, V.; Tri, D.Q.; et al. Fluoride contamination, health problems and remediation methods in Asian groundwater: A comprehensive review. Ecotoxicol. Environ. Saf. 2019, 182, 109362. [Google Scholar] [CrossRef]
- World Health Organization. Guidelines for Drinking-Water Quality: Fourth Edition Incorporating First Addendum; WHO: Geneva, Switzerland, 2017; Available online: https://www.who.int/publications/i/item/9789241549950 (accessed on 10 September 2021).
- Everett, E.T. Fluoride’s effects on the formation of teeth and bones, and the influence of genetics. J. Dent. Res. 2011, 90, 552–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abanto Alvarez, J.; Rezende, K.M.; Marocho, S.M.; Alves, F.B.; Celiberti, P.; Ciamponi, A.L. Dental fluorosis: Exposure, prevention and management. Med. Oral Patol. Oral Cir. Bucal. 2009, 14, 103–107. Available online: http://www.medicinaoral.com/pubmed/medoralv14_i2_pE103.pdf (accessed on 15 September 2021).
- Evans, R.W.; Darvell, B.W. Refining the estimate of the critical period for susceptibility to enamel fluorosis in human maxillary central incisors. J. Public Health Dent. 1995, 55, 238–249. [Google Scholar] [CrossRef]
- Lacruz, R.S.; Habelitz, S.; Wright, J.T.; Paine, M.L. Dental enamel formation and implications for oral health and disease. Physiol. Rev. 2017, 97, 939–993. [Google Scholar] [CrossRef]
- Den Besten, P.K.; Crenshaw, M.A.; Wilson, M.H. Changes in the fluoride-induced modulation of maturation stage ameloblasts of rats. J. Dent. Res. 1985, 64, 1365–1370. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.S.; Wei, X.; Ling, J.Q. Etiology, diagnosis, prevention and treatment of dental fluorosis. Zhonghua Kou Qiang Yi Xue Za Zhi 2020, 55, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Tye, C.E.; Arun, A.; MacDonald, D.; Chatterjee, A.; Abrazinski, T.; Everett, E.T.; Whitford, G.M.; Bartlett, J.D. Assessment of dental fluorosis in Mmp20 ± mice. J. Dent. Res. 2011, 90, 788–792. [Google Scholar] [CrossRef]
- Everett, E.T.; Yin, Z.; Yan, D.; Zou, F. Fine mapping of dental fluorosis quantitative trait loci in mice. Eur. J. Oral Sci. 2011, 119, 8–12. [Google Scholar] [CrossRef] [Green Version]
- Pramanik, S.; Saha, D. The genetic influence in fluorosis. Environ. Toxicol. Pharmacol. 2017, 56, 157–162. [Google Scholar] [CrossRef]
- PROSPERO: International Prospective Register of Systematic Reviews. National Institute for Health Research (NIHR). Available online: https://www.crd.york.ac.uk/prospero/ (accessed on 3 December 2021).
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 29, n71. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.L.; Whaley, P.; Thayer, K.A.; Schünemann, H.J. Identifying the PECO: A framework for formulating good questions to explore the association of environmental and other exposures with health outcomes. Environ. Int. 2018, 121 Pt 1, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Joanna Briggs Institute (JBI). JBI Critical Appraisal Tools (Checklist for Analytical Cross Sectional, Case Control and Cohort Studies). Available online: https://jbi.global/critical-appraisal-tools (accessed on 25 September 2021).
- Higgins, J.P.T.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.; Welch, V. Cochrane Handbook for Systematic Reviews of Interventions Version 6.0 (Updated July 2019). Available online: https://training.cochrane.org/handbook/current (accessed on 2 March 2022).
- McGuinness, L.A.; Higgins, J.P.T. Visualización de riesgo de sesgo (robvis): Un paquete R y una aplicación web Shiny para visualizar evaluaciones de riesgo de sesgo. Res. Synth. Methods 2020, 12, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Ba, Y.; Cui, L.; Cheng, X.; Zhu, J.; Zhang, Y.; Yan, P.; Zhu, C.; Kilfoy, B.; Zhang, Y. COL1A2 gene polymorphisms (Pvu II and Rsa I), serum calciotropic hormone levels, and dental fluorosis. Community Dent. Oral Epidemiol. 2008, 36, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Ba, Y.; Huang, H.; Yang, Y.; Cui, L.; Zhu, J.; Zhu, C.; Liu, J.; Zhang, Y. The association between osteocalcin gene polymorphism and dental fluorosis among children exposed to fluoride in People’s Republic of China. Ecotoxicol. Environ. Saf. 2009, 72, 2158–2161. [Google Scholar] [CrossRef]
- Wang, G.; Ba, Y.; Yang, Y.; Ren, L.; Zhu, J.; Yin, G.; Yu, B.; Cheng, X.; Cui, L. ERβ gene RsaI polymorphism and children’s dental fluorosis. Life Sci. J. 2010, 7, 51–55. Available online: http://www.lifesciencesite.com/lsj/life0701/09_2227_bayue_life0701_51_55.pdf (accessed on 27 September 2021).
- Ba, Y.; Zhang, H.; Wang, G.; Wen, S.; Yang, Y.; Zhu, J.; Ren, L.; Yang, R.; Zhu, C.; Li, H.; et al. Association of dental fluorosis with polymorphisms of estrogen receptor gene in Chinese children. Biol. Trace Element Res. 2011, 143, 87–96. [Google Scholar] [CrossRef]
- Wen, S.; Li, A.; Cui, L.; Huang, Q.; Chen, H.; Guo, X.; Luo, Y.; Hao, Q.; Hou, J.; Ba, Y. The relationship of PTH Bst BI polymorphism, calciotropic hormone levels, and dental fluorosis of children in China. Biol. Trace Element Res. 2012, 147, 84–90. [Google Scholar] [CrossRef]
- Escobar-García, D.; Mejía-Saavedra, J.; Jarquín-Yáñez, L.; Molina-Frechero, N.; Pozos-Guillén, A. Collagenase 1A2 (COL1A2) gene A/C polymorphism in relation to severity of dental fluorosis. Community Dent. Oral Epidemiol. 2016, 44, 162–168. [Google Scholar] [CrossRef]
- Küchler, E.C.; Tannure, P.N.; Oliveira, D.S.; Charone, S.; Nelson-Filho, P.; Silva, R.A.; Costa, M.C.; Antunes, L.S.; Maia, C.; Antunes, L.A. Polymorphisms in genes involved in enamel development are associated with dental fluorosis. Arch. Oral Biol. 2017, 76, 66–69. [Google Scholar] [CrossRef]
- Küchler, E.C.; Dea Bruzamolin, C.; Ayumi Omori, M.; Costa, M.C.; Antunes, L.S.; Pecharki, G.D.; Trevilatto, P.C.; Vieira, A.R.; Brancher, J.A. Polymorphisms in Nonamelogenin Enamel Matrix Genes Are Associated with Dental Fluorosis. Caries Res. 2018, 52, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Jarquín-Yáñez, L.; Alegría-Torres, J.A.; Castillo, C.G.; de Jesús Mejía-Saavedra, J. Dental fluorosis and a polymorphism in the COL1A2 gene in Mexican children. Arch. Oral Biol. 2018, 96, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Dalledone, M.; Cunha, A.S.; Ramazzotto, L.A.; Pecharki, G.D.; Nelson-Filho, P.; Scariot, R.; Trevilatto, P.C.; Vieira, A.R.; Küchler, E.C.; Brancher, J.A. Estrogen receptor gene is associated with dental fluorosis in Brazilian children. Clin. Oral Investig. 2019, 23, 3565–3570. [Google Scholar] [CrossRef] [PubMed]
- Romualdo, P.C.; Pucinelli, C.M.; Tannure, P.N.; Nelson-Filho, P.; Segato, R.A.B.; Brancher, J.A.; Magalhães, N.L.; Costa, M.C.; Antunes, L.A.A.; Antunes, L.S.; et al. Evaluation of genetic polymorphisms in MMP2, MMP9 and MMP20 in Brazilian children with dental fluorosis. Environ. Toxicol. Pharmacol. 2019, 66, 104–108. [Google Scholar] [CrossRef]
- Rahila, C.; Narayanan, A.; Ramesh Kumar, S.G.; Leena Selvamary, A.; Sujatha, A.; John Kirubaharan, J. Association of COL1A2 (PvuII) gene polymorphism with risk and severity of dental fluorosis–A case control study. Saudi Dent. J. 2019, 31, 463–468. [Google Scholar] [CrossRef]
- Tremillo-Maldonado, O.; Molina-Frechero, N.; González-González, R.; Damián-Matsumura, P.; Sánchez-Pérez, L.; Sicco, E.; Suarez, M.; Bologna-Molina, R. DNA sequencing reveals AMELX, ODAM and MMP20 variations in dental fluorosis. Arch. Oral Biol. 2020, 110, 104626. [Google Scholar] [CrossRef]
- Duran-Merino, D.; Molina-Frechero, N.; Sánchez-Pérez, L.; Nevárez-Rascón, M.; González-González, R.; Tremillo-Maldonado, O.; Cassi, D.; Bologna-Molina, R. ENAM Gene Variation in Students Exposed to Different Fluoride Concentrations. Int. J. Environ. Res. Public Health 2020, 17, 1832. [Google Scholar] [CrossRef] [Green Version]
- Abbasoglu, Z.; Dalledone, M.; Wambier, L.M.; Pecharki, G.; Baratto-Filho, F.; Andrades, K.M.R.; Scariot, R.; Trevilatto, P.C.; Brancher, J.A.; Küchler, E.C. Single nucleotide polymorphism rs 4284505 in microRNA17 and risk of dental fluorosis. Acta Odontol. Scand. 2020, 78, 463–466. [Google Scholar] [CrossRef]
- Saha, D.; Goswami, R.; Majumdar, K.K.; Sikdar, N.; Pramanik, S. Evaluating the Association Between Dental Fluorosis and Polymorphisms in Bone Development and Mineralization Genes Among Population from a Fluoride Endemic Region of Eastern India. Biol. Trace Element Res. 2021, 199, 1–8. [Google Scholar] [CrossRef]
- Yuhui, D.; Xiaoli, F.; Jing, J.; Zhiyuan, L.; Kaihong, X.; Meng, G.; Xiangbo, H.; Zichen, F.; Limin, D.; Yongxiang, G.; et al. Effects of SNPs in SOD2 and SOD3 interacted with fluoride exposure on the susceptibility of dental fluorosis. Int. J. Hyg. Environ. Health 2022, 239, 113879. [Google Scholar] [CrossRef]
- Chakraborty, A.; Pramanik, S.; Datta, K.; Goswami, R.; Saha, D.; Majumdar, K.K.; Sikdar, N. Possible Association Between Polymorphisms in ESR1, COL1A2, BGLAP, SPARC, VDR, and MMP2 Genes and Dental Fluorosis in a Population from an Endemic Region of West Bengal. Biol. Trace Element Res. 2022, 200, 4641–4653. [Google Scholar] [CrossRef] [PubMed]
- Piekoszewska-Ziętek, P.; Turska-Szybka, A.; Olczak-Kowalczyk, D. Single Nucleotide Polymorphism in the Aetiology of Caries: Systematic Literature Review. Caries Res. 2017, 51, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Lips, A.; Antunes, L.S.; Antunes, L.A.; Pintor, A.V.B.; Santos, D.A.B.D.; Bachinski, R.; Küchler, E.C.; Alves, G.G. Salivary protein polymorphisms and risk of dental caries: A systematic review. Braz. Oral Res. 2017, 31, e41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almiñana-Pastor, P.J.; Boronat-Catalá, M.; Micó-Martinez, P.; Bellot-Arcís, C.; Lopez-Roldan, A.; Alpiste-Illueca, F.M. Epigenetics and periodontics: A systematic review. Med. Oral Patol. Oral Cir. Bucal. 2019, 24, 659–672. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, Y.J.; Hyun, H.K.; Lee, J.C.; Lee, Z.H.; Kim, J.W. Non-Syndromic Dentinogenesis Imperfecta Caused by Mild Mutations in COL1A2. J. Pers. Med. 2021, 11, 526. [Google Scholar] [CrossRef]
- Riancho, J.A.; Zarrabeitia, M.T.; Valero, C.; Sañudo, C.; Mijares, V.; González-Macías, J. A gene-to-gene interaction between aromatase and estrogen receptors influences bone mineral density. Eur. J. Endocrinol. 2006, 155, 53–59. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W. GSZD. Chronic fluorosis and osteoblastic activation. Chin. J. Control. Endem. Dis. 2003, 18, 210–212. [Google Scholar]
- Ferrer, V.L.; Maeda, T.; Kawano, Y. Characteristic distribution of immunoreaction for estrogen receptor alpha in rat ameloblasts. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2005, 284, 529–536. [Google Scholar] [CrossRef]
- Jedeon, K.; Loiodice, S.; Marciano, C.; Vinel, A.; Canivenc Lavier, M.C.; Berdal, A.; Babajko, S. Estrogen and bisphenol A affect male rat enamel formation and promote ameloblast proliferation. Endocrinology 2014, 155, 3365–3375. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Cui, D.; Zheng, L.; Zhou, Y.; Gan, L.; Liu, Y.; Pan, Y.; Zhou, X.; Wan, M. Bisphenol a Exposure Disrupts Enamel Formation via EZH2-Mediated H3K27me3. J. Dent. Res. 2021, 100, 847–857. [Google Scholar] [CrossRef]
- Bartlett, J.D.; Simmer, J.P. Kallikrein-related peptidase-4 (KLK4): Role in enamel formation and revelations from ablated mice. Front. Physiol. 2014, 5, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, Y.Z.; Mu, L.H.; Wang, Y.X.; An, W.; Jiang, M. Association between ameloblastin gene polymorphisms and the susceptibility to dental fluorosis. Zhonghua Liu Xing Bing Xue Za Zhi 2013, 34, 28–32. [Google Scholar]
- Gerreth, K.; Zaorska, K.; Zabel, M.; Borysewicz-Lewicka, M.; Nowicki, M. Chosen single nucleotide polymorphisms (SNPs) of enamel formation genes and dental caries in a population of Polish children. Adv. Clin. Exp. Med. 2017, 26, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.S.B.; Segato, R.A.B.; Oliveira, S.; Dutra, A.L.T.; Santos, A.S.D.; Praxedes, A.D.N.; Belém, L.C.; Antunes, L.A.; Lips, A.; Nelson-Filho, P.; et al. Association between genetic polymorphisms in DEFB1 and microRNA202 with caries in two groups of Brazilian children. Arch. Oral Biol. 2018, 92, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lézot, F.; Thomas, B.; Greene, S.R.; Hotton, D.; Yuan, Z.A.; Castaneda, B.; Bolaños, A.; Depew, M.; Sharpe, P.; Gibson, C.W.; et al. Physiological implications of DLX homeoproteins in enamel formation. J. Cell. Physiol. 2008, 216, 688–697. [Google Scholar] [CrossRef]
- Li, W.; Jiang, B.; Cao, X.; Xie, Y.; Huang, T. Protective effect of lycopene on fluoride-induced ameloblasts apoptosis and dental fluorosis through oxidative stress-mediated Caspase pathways. Chem. Biol. Interact. 2017, 261, 27–34. [Google Scholar] [CrossRef]




| First Author, Year of Publication | Country | Study Design | Sample | Technique |
|---|---|---|---|---|
| Huang et al. (2008) [18] | China | Case-control | Blood | PCR-RFLP |
| Ba et al. (2009) [19] | China | Case-control | Blood | PCR-RFLP |
| Wang et al. (2010) [20] | China | Case-control | Blood | PCR-RFLP |
| Ba et al. (2011) [21] | China | Case-control | Blood | PCR-RFLP |
| Wen et al. (2012) [22] | China | Case-control | Blood | PCR-RFLP |
| Escobar-García et al. (2016) [23] | Mexico | Cross-sectional | Buccal cells | DNA S |
| Küchler et al. (2017) [24] | Brazil | Cross-sectional | Buccal cells | RT-PCR |
| Küchler et al. (2018) [25] | Brazil | Cohort | Buccal cells | RT-PCR |
| Jarquín-Yáñez et al. (2018) [26] | Mexico | Cross-sectional | Blood | RT-PCR |
| Dalledone et al. (2019) [27] | Brazil | Cross-sectional | Buccal cells | RT-PCR |
| Romualdo et al. (2019) [28] | Brazil | Cross-sectional | Buccal cells | RT-PCR |
| Rahila et al. (2019) [29] | India | Case-control | Blood | PCR-RFLP |
| Tremillo-Maldonado et al. (2020) [30] | Mexico | Cross-sectional | Buccal cells | DNA S |
| Duran-Merino et al. (2020) [31] | Mexico | Cross-sectional | Buccal cells | DNA S |
| Abbasoglu et al. (2020) [32] | Brazil | Cross-sectional | Buccal cells | RT-PCR |
| Saha et al. (2021) [33] | India | Case-control | Blood | PCR-RFLP |
| Yuhui et al. (2022) [34] | China | Cross-sectional | Blood | PCR |
| Chakraborty et al. (2022) [35] | India | Case-control | Blood | PCR–RFLP |
| Reference | Age | Index | N | Cases | Controls |
|---|---|---|---|---|---|
| Huang et al. (2008) [18] | 8–12 | Dean | 240 | 75 | 165 |
| Ba et al. (2009) [19] | 8–12 | Dean | 240 | 75 | 165 |
| Wang et al. (2010) [20] | 8–12 | Dean | 237 | 74 | 163 |
| Ba et al. (2011) [21] | 8–12 | Dean | 240 | 75 | 165 |
| Wen et al. (2012) [22] | 8–12 | Dean | 225 | 68 | 157 |
| Escobar-García et al. (2016) [23] | 6–12 | TF | 80 | 80 | 0 |
| Küchler et al. (2017) [24] | 6–18 | Dean | 481 | 108 | 373 |
| Küchler et al. (2018) [25] | 6–18 | Dean | 1017 | 255 | 762 |
| Jarquín-Yáñez et al. (2018) [26] | 6–12 | TF | 230 | 230 | 0 |
| Dalledone et al. (2019) [27] | 12 | Dean | 538 | 147 | 391 |
| Romualdo et al. (2019) [28] | 6–18 | Dean | 481 | 108 | 373 |
| Rahila et al. (2019) [29] | 10–30 | Dean | 120 | 60 | 60 |
| Tremillo-Maldonado et al. (2020) [30] | NS | TF | 30 | 26 | 4 |
| Duran-Merino et al. (2020) [31] | 11 | TF | 71 | 47 | 24 |
| Abbasoglu et al. (2020) [32] | 10–14 | Dean | 527 | 144 | 383 |
| Saha et al. (2021) [33] | 12–75 | Dean | 87 | 36 | 51 |
| Yuhui et al. (2022) [34] | 7–13 | Dean | 649 | 178 | 471 |
| Chakraborty et al. (2022) [35] | 14–60 | Dean | 132 | 71 | 61 |
| Articles | Answers | Scores | Quality | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |||
| Cross-sectional | |||||||||||||
| Escobar-García et al. (2016) [23] | Y | Y | Y | Y | N | N | Y | Y | -- | -- | -- | 6 | Moderate |
| Küchler et al. (2017) [24] | U | U | Y | Y | Y | Y | Y | Y | -- | -- | -- | 6 | Moderate |
| Jarquín-Yáñez et al. (2018) [26] | Y | Y | Y | Y | N | N | Y | Y | -- | -- | -- | 6 | Moderate |
| Dalledone et al. (2019) [27] | Y | Y | Y | Y | Y | U | Y | Y | -- | -- | -- | 7 | High |
| Romualdo et al. (2019) [28] | U | Y | Y | Y | Y | Y | Y | Y | -- | -- | -- | 7 | High |
| Tremillo-Maldonado et al. (2020) [30] | Y | N | Y | Y | N | N | Y | Y | -- | -- | -- | 5 | Moderate |
| Duran-Merino et al. (2020) [31] | Y | N | Y | Y | N | N | Y | Y | -- | -- | -- | 5 | Moderate |
| Abbasoglu et al. (2020) [32] | Y | Y | Y | Y | N | N | Y | Y | -- | -- | -- | 6 | Moderate |
| Yuhui et al. (2022) [34] | Y | Y | N | Y | Y | Y | Y | Y | -- | -- | -- | 7 | High |
| Case-control | |||||||||||||
| Huang et al. (2008) [18] | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | -- | 9 | High |
| Ba et al. (2009) [19] | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | -- | 9 | High |
| Wang et al. (2010) [20] | Y | Y | Y | U | Y | Y | Y | Y | Y | Y | -- | 9 | High |
| Ba et al. (2011) [21] | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | -- | 10 | High |
| Wen et al. (2012) [22] | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | -- | 9 | High |
| Rahila et al. (2019) [29] | Y | Y | Y | N | Y | Y | U | Y | Y | Y | -- | 8 | Moderate |
| Saha et al. (2022) [33] | Y | Y | Y | N | Y | Y | U | N | Y | Y | -- | 7 | Moderate |
| Chakraborty et al. (2022) [35] | Y | Y | Y | N | Y | Y | U | N | Y | Y | -- | 7 | Moderate |
| Cohort | |||||||||||||
| Küchler et al. (2018) [25] | U | Y | Y | Y | Y | Y | Y | N | N | N | Y | 7 | Moderate |
| Author | Gene: SNP | Results |
|---|---|---|
| Huang et al. (2008) [18] | COL1A2: rs414408 | P allele increased the risk of DF (OR = 4.85, IC95%: 1.22–19.32). |
| Ba et al. (2011) [21] | ESR2: rs1256049 | R allele increased the risk of DF (OR = 1.821; IC95%: 1.013–3.274). |
| Küchler et al. (2017) [24] | DLX1: rs788173 DLX2: rs743605 | In both SNPs, AG genotypes were associated with the most severe form of DF (p < 0.05). |
| Küchler et al. (2018) [25] | AMBN: rs4694075 | T allele increased the risk of DF (p < 0.0001; OR = 2.12, 95% CI: 1.51–2.97) |
| TUFT1: rs4970957 | A allele increased the risk of DF (p < 0.049; OR = 1.42, 95% CI: 1.00–2.02). | |
| TFIP11: rs5997096 | T allele was associated with the moderate/severe form of DF (p = 0.005). | |
| Jarquín-Yáñez et al. (2018) [26] | COL1A2: rs412777 | C allele increased the risk of DF (p = 0.05, OR = 2.59, IC95%: 1.60–4.20) |
| Rahila et al. (2019) [29] | COL1A2: rs414408 | PP and Pp genotypes increased the risk of DF (OR = 31.4; IC95%: 3.9–48.7 and OR = 4.0, IC95%: 1.6–10.1) |
| Abbasoglu et al. (2020) [31] | miARN17:rs4284505 | G allele increased the risk of DF (p = 0.031; OR = 2.26, IC95%: 1.04–4.73) |
| Yuhui et al. (2022) [34] | SOD2: rs10370 | G allele increased the risk of DF (IC95% for OR: 1.20, 2.96) |
| SOD2: rs5746136 | T allele increased the risk of DF (IC95% for OR: 1.09, 2.69) |
| Author | Gene: SNP | Results |
|---|---|---|
| Ba et al. (2011) [21] | ESR1: rs2234693 | X allele decreased the risk of DF (OR = 0.542, IC95%: 0.314–0.936). |
| Dalledone et al. (2019) [27] | ESR1: rs12154178 | CC genotype decreased the risk of DF (p = 0.038; OR = 0.51, IC95%: 0.7–0.97). |
| Tremillo-Maldonado et al. (2020) [30] | MMP20: rs1784418 | TG allele variant decreased the risk of DF (p = 0.001). |
| Duran-Merino et al. (2020) [31] | ENAM: rs12640848 | AG/CT allele variant was associated with a lower severity of DF (p = 0.000). |
| Author | Gene: SNP | Results |
|---|---|---|
| Küchler et al. (2017) [24] | TIMP1: rs4898 | Borderline association with DF was observed in CT genotype (p = 0.073). |
| Romualdo et al. (2019) [28] | Borderline association with DF was observed in: | |
| MMP2: rs243865 | CT genotype (p = 0.06). | |
| MMP9: rs17576 | AG genotype (p = 0.08). | |
| MMP20: rs1784418 | AG genotype (p= 0.06). |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Casamada, C.; Nevarez-Rascón, M.; Nevarez-Rascón, A.; González-Galván, M.; Isiordia-Espinoza, M.A.; Bologna-Molina, R.; Sánchez-Pérez, L.; Molina-Frechero, N. Single Nucleotide Polymorphisms and Dental Fluorosis: A Systematic Review. Dent. J. 2022, 10, 211. https://doi.org/10.3390/dj10110211
González-Casamada C, Nevarez-Rascón M, Nevarez-Rascón A, González-Galván M, Isiordia-Espinoza MA, Bologna-Molina R, Sánchez-Pérez L, Molina-Frechero N. Single Nucleotide Polymorphisms and Dental Fluorosis: A Systematic Review. Dentistry Journal. 2022; 10(11):211. https://doi.org/10.3390/dj10110211
Chicago/Turabian StyleGonzález-Casamada, Carlos, Martina Nevarez-Rascón, Alfredo Nevarez-Rascón, María González-Galván, Mario Alberto Isiordia-Espinoza, Ronell Bologna-Molina, Leonor Sánchez-Pérez, and Nelly Molina-Frechero. 2022. "Single Nucleotide Polymorphisms and Dental Fluorosis: A Systematic Review" Dentistry Journal 10, no. 11: 211. https://doi.org/10.3390/dj10110211
APA StyleGonzález-Casamada, C., Nevarez-Rascón, M., Nevarez-Rascón, A., González-Galván, M., Isiordia-Espinoza, M. A., Bologna-Molina, R., Sánchez-Pérez, L., & Molina-Frechero, N. (2022). Single Nucleotide Polymorphisms and Dental Fluorosis: A Systematic Review. Dentistry Journal, 10(11), 211. https://doi.org/10.3390/dj10110211