The Potential Clinical Applications of a Microfluidic Lab-on-a-Chip for the Identification and Antibiotic Susceptibility Testing of Enterococcus faecalis-Associated Endodontic Infections: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Approach
2.2. Selection Criteria
2.3. Question
2.4. Review Course
2.5. Compilation of Data
2.6. Risk of Bias
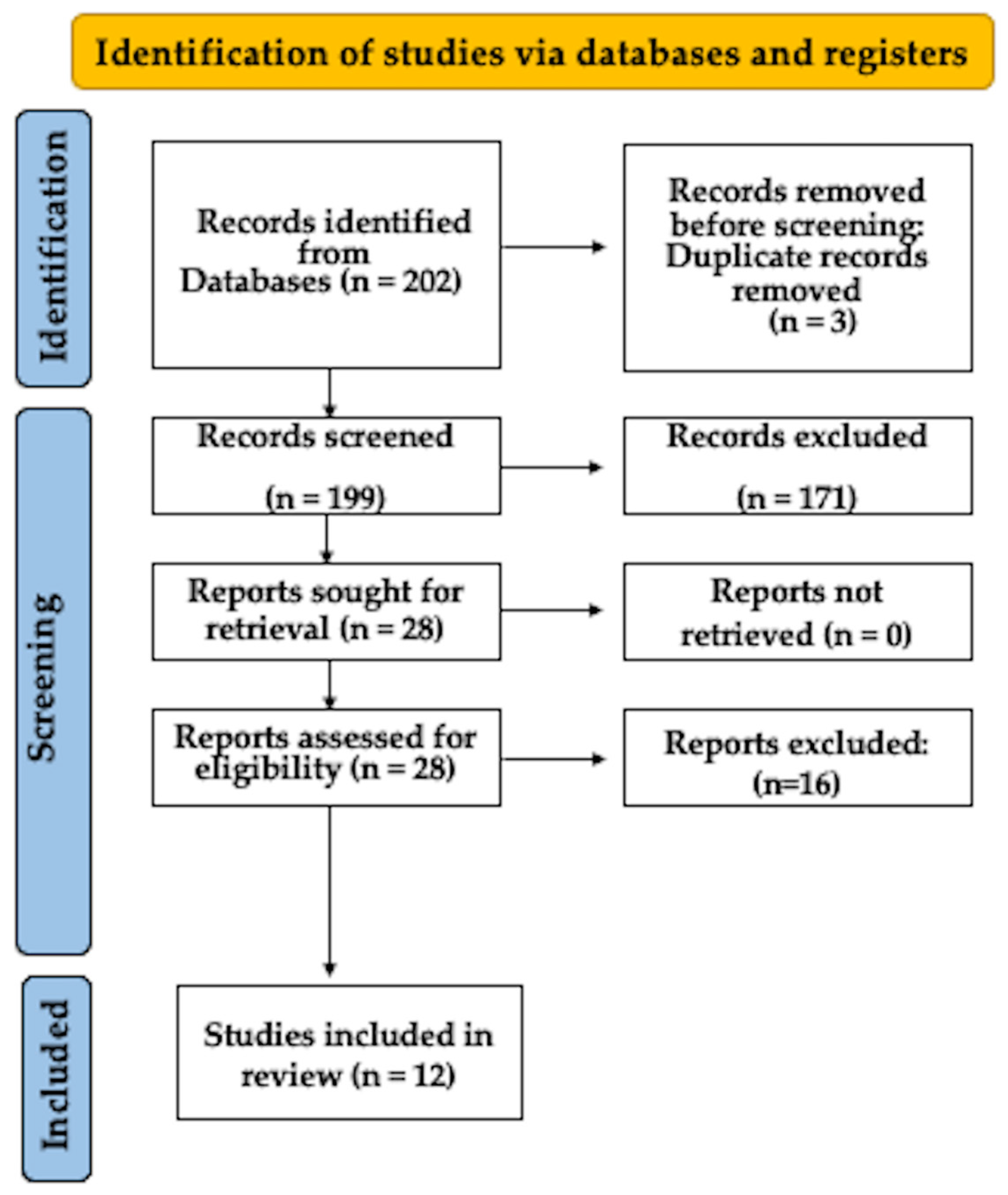
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [PubMed]
- Elashiry, M.M.; Bergeron, B.E.; Tay, F.R. Enterococcus faecalis in secondary apical periodontitis: Mechanisms of bacterial survival and disease persistence. Microb. Pathog. 2023, 183, 106337. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.A.A.; Cheung, G.S.P.; Neelakantan, P. Transition metals and Enterococcus faecalis: Homeostasis, virulence, and perspectives. Mol. Oral Microbiol. 2022, 37, 276–291. [Google Scholar] [CrossRef] [PubMed]
- Manoil, D.; Al-Manei, K.; Belibasakis, G.N. A Systematic Review of the Root Canal Microbiota Associated with Apical Periodontitis: Lessons from Next-Generation Sequencing. Proteom. Clin. Appl. 2020, 14, e1900060. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Manoil, D.; Näsman, P.; Belibasakis, G.N.; Neelakantan, P. Microbiological Aspects of Root Canal Infections and Disinfection Strategies: An Update Review on the Current Knowledge and Challenges. Front. Oral Health 2021, 2, 672887. [Google Scholar] [CrossRef]
- Gaeta, C.; Marruganti, C.; Ali, I.A.A.; Fabbro, A.; Pinzauti, D.; Santoro, F.; Neelakantan, P.; Pozzi, G.; Grandini, S. The presence of Enterococcus faecalis in saliva as a risk factor for endodontic infection. Front. Cell. Infect. Microbiol. 2023, 13, 1061645. [Google Scholar] [CrossRef]
- Momenijavid, M.; Salimizand, H.; Korani, A.; Dianat, O.; Nouri, B.; Ramazanzadeh, R.; Ahmadi, A.; Rostamipour, J.; Khosravi, M.R. Effect of calcium hydroxide on morphology and physicochemical properties of Enterococcus faecalis biofilm. Sci. Rep. 2022, 12, 7595. [Google Scholar] [CrossRef]
- Gomes, B.P.F.A.; Francisco, P.A.; Godoi, E.P., Jr.; Endo, M.S.; Barbosa-Ribeiro, M.; Delboni, M.G.; Pecorari, V.G.A. Identification of Culturable and Nonculturable Microorganisms, Lipopolysaccharides, and Lipoteichoic Acids from Root Canals of Teeth with Endodontic Failure. J. Endod. 2021, 47, 1075–1086. [Google Scholar] [CrossRef]
- Delboni, M.G.; Gomes, B.P.; Francisco, P.A.; Teixeira, F.B.; Drake, D. Diversity of Enterococcus faecalis Genotypes from Multiple Oral Sites Associated with Endodontic Failure Using Repetitive Sequence-based Polymerase Chain Reaction and Arbitrarily Primed Polymerase Chain Reaction. J. Endod. 2017, 43, 377–382. [Google Scholar] [CrossRef]
- Vianna, M.E.; Horz, H.P.; Gomes, B.P.; Conrads, G. Microarrays complement culture methods for identification of bacteria in endodontic infections. Oral Microbiol. Immunol. 2005, 20, 253–258. [Google Scholar] [CrossRef]
- Barbosa-Ribeiro, M.; Arruda-Vasconcelos, R.; Louzada, L.M.; Dos Santos, D.G.; Andreote, F.D.; Gomes, B.P.F.A. Microbiological analysis of endodontically treated teeth with apical periodontitis before and after endodontic retreatment. Clin. Oral Investig. 2021, 25, 2017–2027. [Google Scholar] [CrossRef] [PubMed]
- Nóbrega, L.M.; Montagner, F.; Ribeiro, A.C.; Mayer, M.A.; Gomes, B.P. Bacterial diversity of symptomatic primary endodontic infection by clonal analysis. Braz. Oral Res. 2016, 30, e103. [Google Scholar] [CrossRef] [PubMed]
- Welker, M.; van Belkum, A. One System for All: Is Mass Spectrometry a Future Alternative for Conventional Antibiotic Susceptibility Testing? Front. Microbiol. 2019, 10, 2711. [Google Scholar] [CrossRef] [PubMed]
- Arabestani, M.R.; Rastiany, S.; Kazemi, S.; Mousavi, S.M. Conventional, molecular methods and biomarkers molecules in detection of septicemia. Adv. Biomed. Res. 2015, 4, 120. [Google Scholar] [CrossRef] [PubMed]
- Ardila, C.M.; Jiménez-Arbeláez, G.A.; Vivares-Builes, A.M. Potential Clinical Application of Organs-on-a-Chip in Periodontal Diseases: A Systematic Review of In Vitro Studies. Dent. J. 2023, 11, 158. [Google Scholar] [CrossRef] [PubMed]
- Ardila, C.M.; Zuluaga-Gómez, M.; Vivares-Builes, A.M. Applications of Lab on a Chip in Antimicrobial Susceptibility of Staphylococcus aureus: A Systematic Review. Medicina 2023, 59, 1719. [Google Scholar] [CrossRef]
- Tong, Z.; Shen, C.; Li, Q.; Yin, H.; Mao, H. Combining sensors and actuators with electrowetting-on-dielectric (EWOD): Advanced digital microfluidic systems for biomedical applications. Analyst 2023, 148, 1399–1421. [Google Scholar] [CrossRef]
- Ma, X.; Guo, G.; Wu, X.; Wu, Q.; Liu, F.; Zhang, H.; Shi, N.; Guan, Y. Advances in Integration, Wearable Applications, and Artificial Intelligence of Biomedical Microfluidics Systems. Micromachines 2023, 14, 972. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Sheth, V.H.; Shah, N.P.; Jain, R.; Bhanushali, N.; Bhatnagar, V. Development and validation of a risk-of-bias tool for assessing in vitro studies conducted in dentistry: The QUIN. J. Prosthet. Dent. 2022. [Google Scholar] [CrossRef]
- Mahalanabis, M.; Al-Muayad, H.; Kulinski, M.D.; Altman, D.; Klapperich, C.M. Cell lysis and DNA extraction of gram-positive and gram-negative bacteria from whole blood in a disposable microfluidic chip. Lab Chip 2009, 9, 2811–2817. [Google Scholar] [CrossRef] [PubMed]
- Cira, N.J.; Ho, J.Y.; Dueck, M.E.; Weibel, D.B. A self-loading microfluidic device for determining the minimum inhibitory concentration of antibiotics. Lab Chip 2012, 12, 1052–1059. [Google Scholar] [CrossRef] [PubMed]
- Puchberger-Enengl, D.; van den Driesche, S.; Krutzler, C.; Keplinger, F.; Vellekoop, M.J. Hydrogel-based microfluidic incubator for microorganism cultivation and analyses. Biomicrofluidics 2015, 9, 014127. [Google Scholar] [CrossRef] [PubMed]
- Schröder, U.C.; Bokeloh, F.; O′Sullivan, M.; Glaser, U.; Wolf, K.; Pfister, W.; Popp, J.; Ducrée, J.; Neugebauer, U. Rapid, culture-independent, optical diagnostics of centrifugally captured bacteria from urine samples. Biomicrofluidics 2015, 9, 044118. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, R.; Chen, J.L.; Stuckey, D.C.; Steele, T.W.J. Poly(methyl methacrylate) Surface Modification for Surfactant-Free Real-Time Toxicity Assay on Droplet Microfluidic Platform. ACS Appl. Mater. Interfaces 2017, 9, 13801–13811. [Google Scholar] [CrossRef] [PubMed]
- Azizi, M.; Zaferani, M.; Dogan, B.; Zhang, S.; Simpson, K.W.; Abbaspourrad, A. Nanoliter-Sized Microchamber/Microarray Microfluidic Platform for Antibiotic Susceptibility Testing. Anal. Chem. 2018, 90, 14137–14144. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.Y.; Cira, N.J.; Crooks, J.A.; Baeza, J.; Weibel, D.B. Rapid identification of ESKAPE bacterial strains using an autonomous microfluidic device. PLoS ONE 2012, 7, e41245. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Sarkar, S.; Lin, Z.S.; McKenney, S.; Konry, T. Ultrafast Parallelized Microfluidic Platform for Antimicrobial Susceptibility Testing of Gram Positive and Negative Bacteria. Anal. Chem. 2019, 91, 6242–6249. [Google Scholar] [CrossRef]
- Dreier, M.; Berthoud, H.; Shani, N.; Wechsler, D.; Junier, P. SpeciesPrimer: A bioinformatics pipeline dedicated to the design of qPCR primers for the quantification of bacterial species. PeerJ 2020, 8, e8544. [Google Scholar] [CrossRef]
- Kao, Y.T.; Kaminski, T.S.; Postek, W.; Guzowski, J.; Makuch, K.; Ruszczak, A.; von Stetten, F.; Zengerle, R.; Garstecki, P. Gravity-driven microfluidic assay for digital enumeration of bacteria and for antibiotic susceptibility testing. Lab Chip 2020, 20, 54–63. [Google Scholar] [CrossRef]
- Gowda, H.N.; Kido, H.; Wu, X.; Shoval, O.; Lee, A.; Lorenzana, A.; Madou, M.; Hoffmann, M.; Jiang, S.C. Development of a proof-of-concept microfluidic portable pathogen analysis system for water quality monitoring. Sci. Total Environ. 2022, 813, 152556. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Jia, Y.; Li, R.; Wen, Y.; Liang, Y.; Lao, G.; Liu, X.; Zhou, W.; Liu, H.; Xie, J.; et al. Rapid and simultaneous detection of multiple pathogens in the lower reproductive tract during pregnancy based on loop-mediated isothermal amplification-microfluidic chip. BMC Microbiol. 2022, 22, 260. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; CLSI Document M07-A11; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Desmet, S.; Verhaegen, J.; Glupzcynski, Y.; Van Eldere, J.; Melin, P.; Goossens, H.; Piérard, D.; Declercq, P.; Lagrou, K.; Boel, A.; et al. Development of a national EUCAST challenge panel for antimicrobial susceptibility testing. Clin. Microbiol. Infect. 2016, 22, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, A.; Harris, D.; Selvaganapathy, P.R.; Kishen, A. Electrokinetic transport and distribution of antibacterial nanoparticles for endodontic disinfection. Int. Endod. J. 2020, 53, 1120–1130. [Google Scholar] [CrossRef]
- França, C.M.; Tahayeri, A.; Rodrigues, N.S.; Ferdosian, S.; Puppin Rontani, R.M.; Sereda, G.; Ferracane, J.L.; Bertassoni, L.E. The tooth on-a-chip: A microphysiologic model system mimicking the biologic interface of the tooth with biomaterials. Lab Chip 2020, 20, 405–413. [Google Scholar] [CrossRef]
- Rodrigues, N.S.; França, C.M.; Tahayeri, A.; Ren, Z.; Saboia, V.P.A.; Smith, A.J.; Ferracane, J.L.; Koo, H.; Bertassoni, L.E. Biomaterial and Biofilm Interactions with the Pulp-Dentin Complex-on-a-Chip. J. Dent. Res. 2021, 100, 1136–1143. [Google Scholar] [CrossRef]
- Cheng, X.; Tian, Y.; Zhao, C.; Qu, T.; Ma, C.; Liu, X.; Yu, Q. Bactericidal Effect of Strong Acid Electrolyzed Water against Flow Enterococcus faecalis Biofilms. J. Endod. 2016, 42, 1120–1125. [Google Scholar] [CrossRef]
- Ahrberg, C.D.; Manz, A.; Chung, B.G. Polymerase chain reaction in microfluidic devices. Lab Chip 2016, 16, 3866–3884. [Google Scholar] [CrossRef]
- Li, J.; Zhu, Y.; Wu, X.; Hoffmann, M.R. Rapid Detection Methods for Bacterial Pathogens in Ambient Waters at the Point of Sample Collection: A Brief Review. Clin. Infect. Dis. 2020, 71 (Suppl. S2), S84–S90. [Google Scholar] [CrossRef]
- Scheler, O.; Pacocha, N.; Debski, P.R.; Ruszczak, A.; Kaminski, T.S.; Garstecki, P. Optimized droplet digital CFU assay (ddCFU) provides precise quantification of bacteria over a dynamic range of 6 logs and beyond. Lab Chip 2017, 17, 1980–1987. [Google Scholar] [CrossRef]
- Jiang, L.; Boitard, L.; Broyer, P.; Chareire, A.C.; Bourne-Branchu, P.; Mahé, P.; Tournoud, M.; Franceschi, C.; Zambardi, G.; Baudry, J.; et al. Digital antimicrobial susceptibility testing using the MilliDrop technology. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Baltekin, Ö.; Boucharin, A.; Tano, E.; Andersson, D.I.; Elf, J. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc. Natl. Acad. Sci. USA 2017, 114, 9170–9175. [Google Scholar] [CrossRef] [PubMed]
- Veses-Garcia, M.; Antypas, H.; Löffler, S.; Brauner, A.; Andersson-Svahn, H.; Richter-Dahlfors, A. Rapid Phenotypic Antibiotic Susceptibility Testing of Uropathogens Using Optical Signal Analysis on the Nanowell Slide. Front. Microbiol. 2018, 9, 1530. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, S.; García-Aznar, J.M.; Gonzalo-Asensio, J. Microfluidic devices for studying bacterial taxis, drug testing and biofilm formation. Microb. Biotechnol. 2022, 15, 395–414. [Google Scholar] [CrossRef]
- Huang, H.K.; Cheng, H.W.; Liao, C.C.; Lin, S.J.; Chen, Y.Z.; Wang, J.K.; Wang, Y.L.; Huang, N.T. Bacteria encapsulation and rapid antibiotic susceptibility test using a microfluidic microwell device integrating surface-enhanced Raman scattering. Lab Chip 2020, 20, 2520–2528. [Google Scholar] [CrossRef]
- Wistrand-Yuen, P.; Malmberg, C.; Fatsis-Kavalopoulos, N.; Lübke, M.; Tängdén, T.; Kreuger, J. A Multiplex Fluidic Chip for Rapid Phenotypic Antibiotic Susceptibility Testing. mBio 2020, 11, e03109-19. [Google Scholar] [CrossRef]
| Materials Used | Culture, Bacterial Strains, and Growing Conditions | Main Results | Reference |
|---|---|---|---|
| The microchannels were generated through the process of hot embossing, utilizing a nickel–cobalt electroplated mould that was fabricated based on a silicon master template. | The PCR amplification of the extracted DNA was evaluated through the introduction of E. faecalis (ATCC 29212) into. E. faecalis was cultivated in 3 mL of brain heart infusion media at a temperature of 37 degrees Celsius for a duration of 16 to 18 h, with agitation at 250 revolutions per minute. | The genomic DNA of E. faecalis was successfully isolated and identified in microlitre-scale human whole-blood samples. The entire extraction process required less than an hour and can be further reduced by modifying the channel design and the pump setup. | [21] |
| The devices were manufactured employing soft lithography techniques. Specifically, masters were created by applying SU-8 photoresist (Microchem) onto silicon wafers through the process of photolithography. | Off-chip minimum inhibitory concentration assays were conducted in accordance with the Clinical and Laboratory Standards Institute protocols. Cell suspensions of E. faecalis (strain 1131) were prepared in 13 × 100 mm tubes by adjusting the cell concentration from overnight cultures to match the turbidity of a 0.5 McFarland standard, approximately ~108 CFU/mL, using cation-adjusted Mueller–Hinton broth. | Through the utilization of a microfluidic device, the MIC of vancomycin, tetracycline, and kanamycin against E. faecalis was determined, and the obtained MIC values were found to be consistent with those derived from conventional liquid broth dilution techniques. Notably, it was observed that bacterial cultures within the microfluidic devices achieved their MIC values more rapidly when compared to off-chip assays. | [22] |
| The microfluidic devices were made via hot roll lamination, which allows for quick, parallel, and cost-effective production. A colorimetric colour change is the simplest approach to confirm bacterial growth in a microfluidic chip. | The gel was produced with 20 mg/mL lysogenic broth for bacterial testing. On lysogenic broth agar plates, E. faecalis (DSM 16440) was grown. A colony was diluted in 10 mM phosphate-buffered saline to make the samples. | Ampicillin >1 µg/mL inhibited E. faecalis growth and Gentamicin resistance was observed at all doses. The quick microfluidic method reliably assesses susceptibility with MIC concentrations in agreement with standard reference methods. | [23] |
| This platform employs the centrifugal force generated by rotation to effectively trap bacteria directly from a liquid suspension within a chip. This is a polymeric microfluidic system. | E. faecalis ATCC 29212 was grown on agar at 37 °C for one night. | The entire process, from sample preparation to obtaining valuable results, takes approximately 1 h. This represents a substantial reduction in diagnostic time compared to the typically lengthier period of 24 h or more required for standard microbiological methods. | [24] |
| A layer of silica nanoparticles was applied to a surface made of polymethylmethacrylate and other thermoplastics. | E. faecalis (ATCC 47077) was introduced into 10 mL of sterile brain heart infusion medium that had been purged with nitrogen. The culture was allowed to incubate overnight, reaching the stationary phase, and subsequently stored at 4 °C for use within 24 h. On the microchip, droplets containing nickel cations, combined with bacteria and resazurin, were created. | The detection of metabolic inhibition in E. faecalis occurred within a 5 min timeframe. The entire procedure, which included the sequential injection of reagents and the simultaneous monitoring of droplet fluorescence intensity, was conducted directly on the microchip. | [25] |
| The silicon wafer was imprinted with mould using the established soft lithography method. | E. faecalis 24 was agitated and cultured in a brain heart infusion (BHI) medium for 12 h at 37 °C within a 2% BHI broth environment. An imaging platform was employed to measure the fluorescence intensity generated in the bacterial culture medium because of the redox reaction involving resazurin. | The microfluidic platform exhibited quicker performance, completing its task in 1 to 3 h, as opposed to the conventional gold standard, broth microdilution, which typically takes 12 to 18 h. Despite the speed, it maintained a similar level of accuracy. Ampicillin, kanamycin, and gentamicin were all effective against E. faecalis. | [26] |
| Masters were created in SU-8 photoresist (Microchem) on silicon wafers through the process of photolithography. | For E. faecalis identification, the bacteria were cultured at 37 °C overnight on Luria–Bertani (LB) agar. Afterward, isolated and selected individual colonies were transferred to a 100 mL solution of 25% LB. It homogenized the inoculum through vortexing and subsequently introduced 20 mL of cell suspension into a BacChip. | This automated microfluidic system can identify E. faecalis within a timeframe of less than 4 h. | [27] |
| The production of the microfluidic device primarily comprises the photolithography process for creating the SU-8 master template, soft lithography to produce polydimethylsiloxane replicas, and subsequently, bonding the device to microscope slides through plasma treatment. | E. faecalis (ATCC 29212) was cultivated in brain heart infusion agar/broth, following the procedures specified by ATCC. The growth of the bacteria was observed in a minimum of 100 droplets, utilizing time-lapse imaging, over a period of 2 h, with images captured at 15 min intervals. | The MIC derived through phenotypic analysis within droplets correlated with the MIC results obtained via the conventional broth microdilution method. Nonetheless, this method is notably swifter (30 min versus 16 to 24 h). All oxacillin concentrations greatly suppressed E. faecalis growth. | [28] |
| A microfluidic-based high-throughput qPCR assay was created. The entire primer design process, from fetching bacterial genome data to vetting primer candidates for quality, was automated. | Strains preserved in the Agroscope Culture Collection at −80 °C within sterile skim milk powder were reawakened and grown as per previously defined conditions. | The SpeciesPrimer pipeline was finalized within a timeframe of two to eight hours. SpeciesPrimer streamlines the process of primer design for precise species quantification, enabling a swift and precise quantitative analysis of E. faecalis. | [29] |
| Polydimethylsiloxane was poured over a polycarbonate master created through CNC milling and then incubated at 70 °C for a duration of 2 h. | E. faecalis (ATCC 51299) was cultured on agar plates containing 2% MH broth and then incubated at 37 °C overnight. | E. faecalis was susceptible to ampicillin. It was possible to significantly cut down the assay time to approximately 5 h, a notable improvement compared to the 20 h required by the conventional culture-based test. | [30] |
| The microfluidic centrifugal disc comprises four fundamental layers constructed from polycarbonate. | E. faecalis (ATCC 29212) was grown overnight in Luria–Bertani broth at 37 °C. Following that, the culture was serially diluted in deionized water and used for loop-mediated isothermal amplification, either in tubes or on a centrifuge disc. | The prototype device can detect E. faecalis in water samples by just pressing a start button, and the process takes 1 h with a total hands-on time of less than 5 min. | [31] |
| The LAMP-microfluidic chip was created using a mix of loop-mediated isothermal amplification (LAMP) and microfluidic technology. | Lyophilized standard strains (E. faecalis CGMCC 1.10682) and cryopreserved clinical isolates were cultured on appropriate agar plates before being picked for rejuvenation. Three repeated detections on three random clinical samples with positive results identified by LAMP-microfluidic chip were performed. | The disclosed LAMP-microfluidic chip approach can identify E. faecalis quickly, and the entire procedure from DNA extraction to amplification completion took just about 90 min, with detection sensitivities of less than 105 CFU/mL or copies/mL. | [32] |
| * Criteria Met | ^ Points | + Score | Reference |
|---|---|---|---|
| 8 | 14 | 58% | [21] |
| 8 | 14 | 58% | [22] |
| 8 | 14 | 58% | [23] |
| 8 | 14 | 58% | [24] |
| 8 | 14 | 58% | [25] |
| 8 | 14 | 58% | [26] |
| 8 | 14 | 58% | [27] |
| 8 | 14 | 58% | [28] |
| 8 | 14 | 58% | [29] |
| 8 | 14 | 58% | [30] |
| 9 | 16 | 67% | [31] |
| 9 | 16 | 67% | [32] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ardila, C.M.; Jiménez-Arbeláez, G.A.; Vivares-Builes, A.M. The Potential Clinical Applications of a Microfluidic Lab-on-a-Chip for the Identification and Antibiotic Susceptibility Testing of Enterococcus faecalis-Associated Endodontic Infections: A Systematic Review. Dent. J. 2024, 12, 5. https://doi.org/10.3390/dj12010005
Ardila CM, Jiménez-Arbeláez GA, Vivares-Builes AM. The Potential Clinical Applications of a Microfluidic Lab-on-a-Chip for the Identification and Antibiotic Susceptibility Testing of Enterococcus faecalis-Associated Endodontic Infections: A Systematic Review. Dentistry Journal. 2024; 12(1):5. https://doi.org/10.3390/dj12010005
Chicago/Turabian StyleArdila, Carlos M., Gustavo A. Jiménez-Arbeláez, and Annie Marcela Vivares-Builes. 2024. "The Potential Clinical Applications of a Microfluidic Lab-on-a-Chip for the Identification and Antibiotic Susceptibility Testing of Enterococcus faecalis-Associated Endodontic Infections: A Systematic Review" Dentistry Journal 12, no. 1: 5. https://doi.org/10.3390/dj12010005
APA StyleArdila, C. M., Jiménez-Arbeláez, G. A., & Vivares-Builes, A. M. (2024). The Potential Clinical Applications of a Microfluidic Lab-on-a-Chip for the Identification and Antibiotic Susceptibility Testing of Enterococcus faecalis-Associated Endodontic Infections: A Systematic Review. Dentistry Journal, 12(1), 5. https://doi.org/10.3390/dj12010005










