A Docking and Network Pharmacology Study on the Molecular Mechanisms of Curcumin in Dental Caries and Streptococcus mutans
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. GO and Pathway Enrichment Analyses
2.3. Construction of the Target-Curcumin-Metabolic Pathways Network in Caries
2.4. Molecular Docking
2.5. Effect of Curcumin on Streptococcus mutans
3. Results
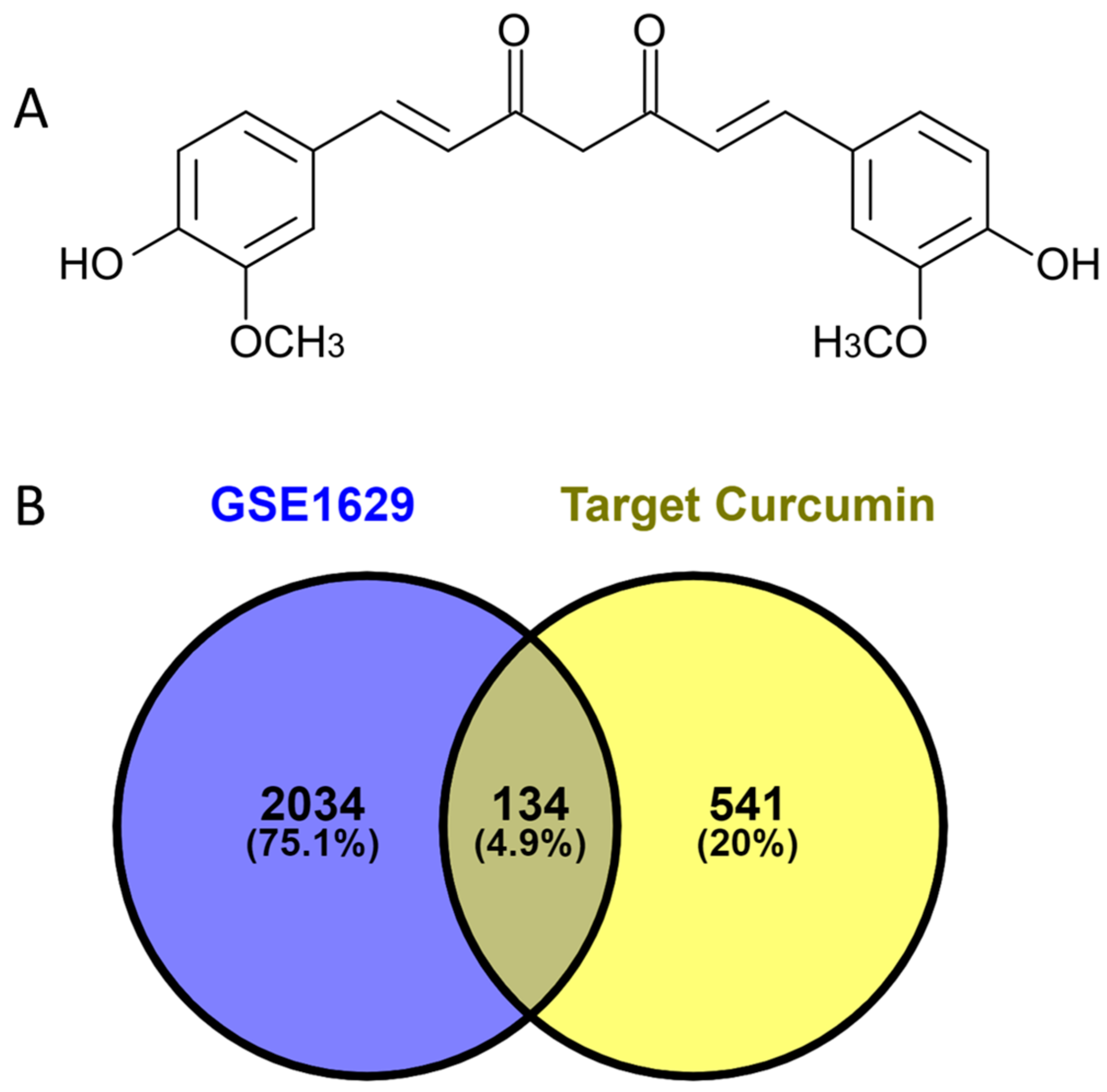
3.1. Analysis of Curcumin Targets and Caries
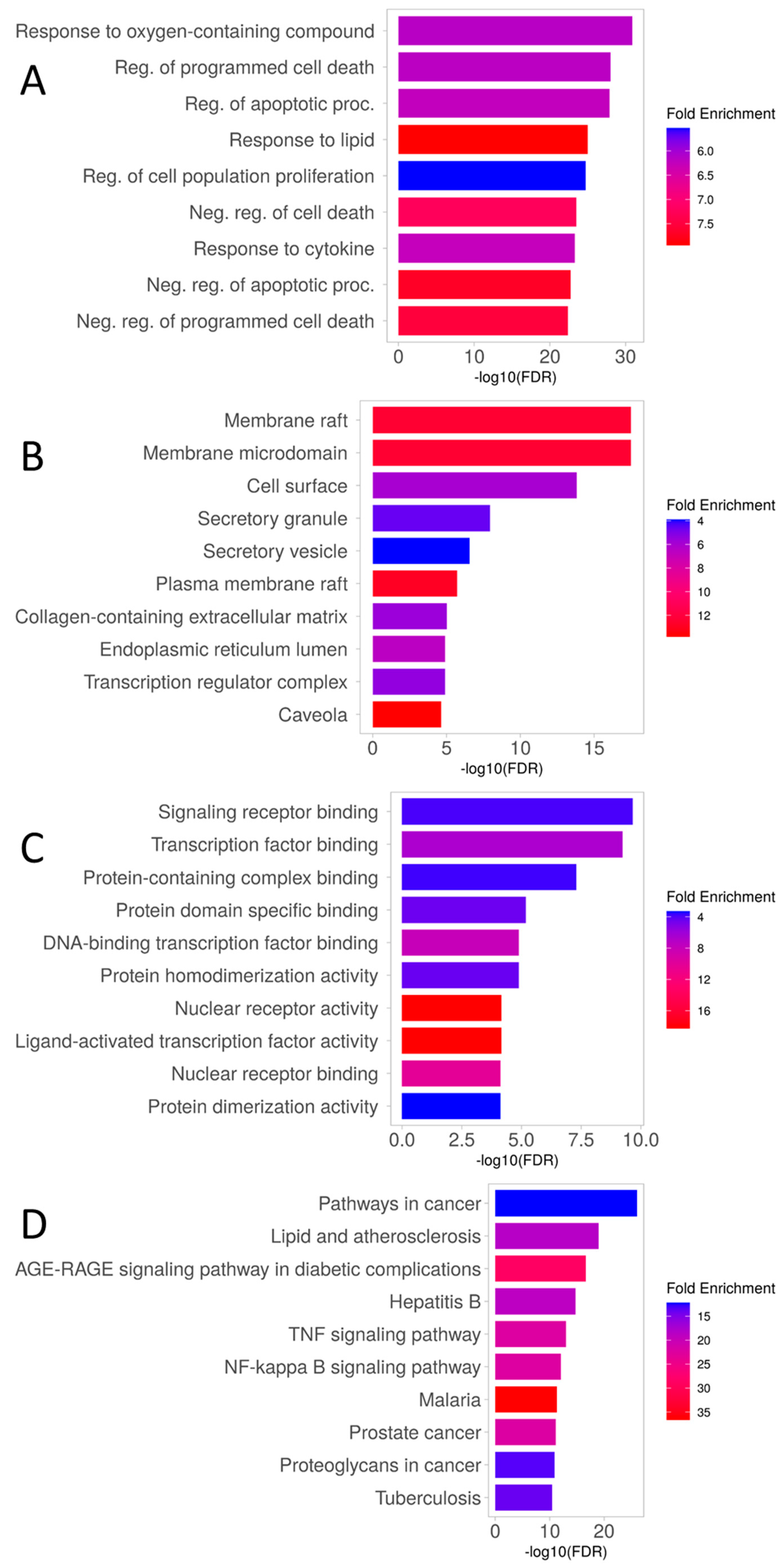
3.2. Enrichment Analysis of Overlapping Targets
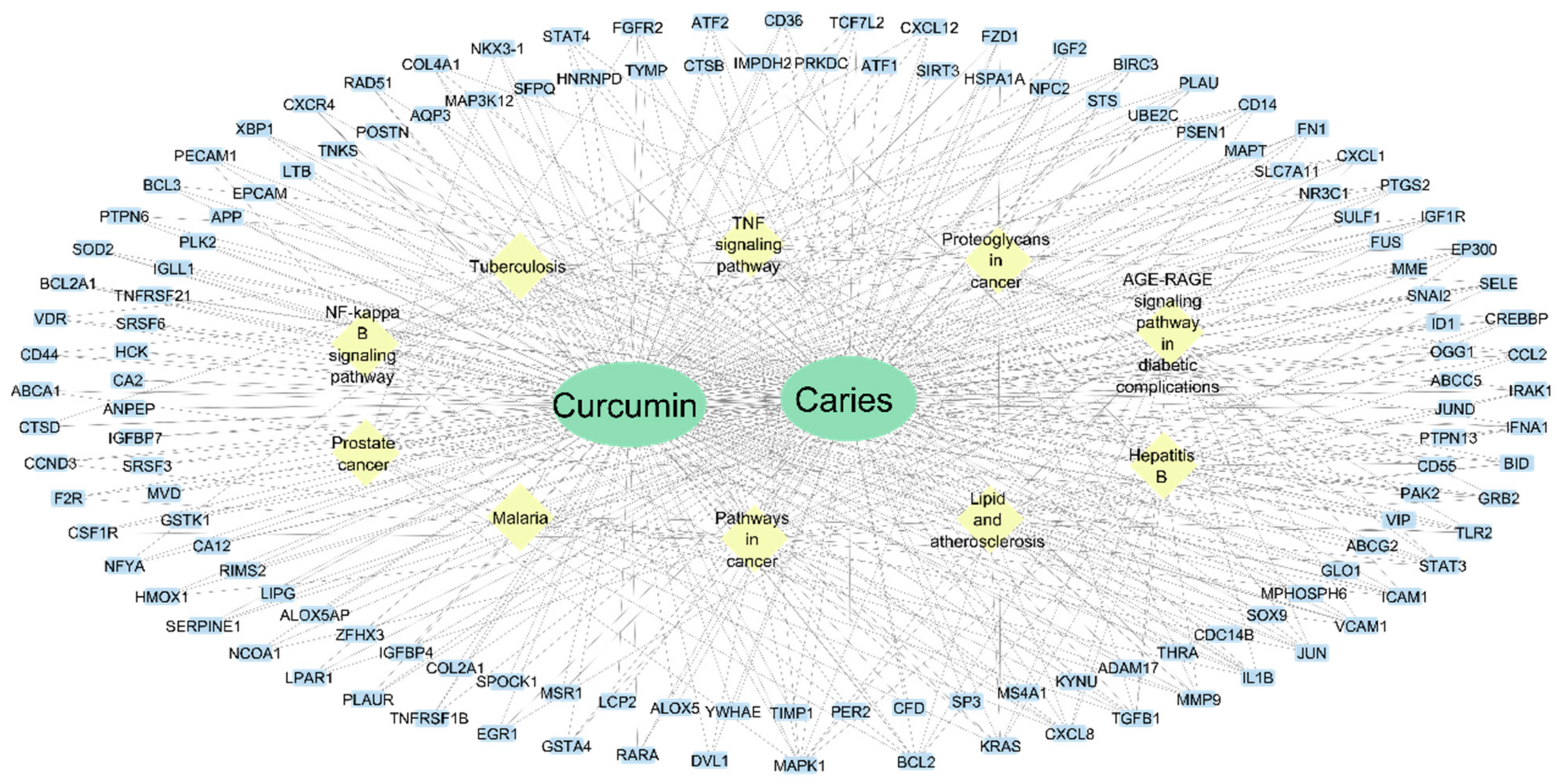
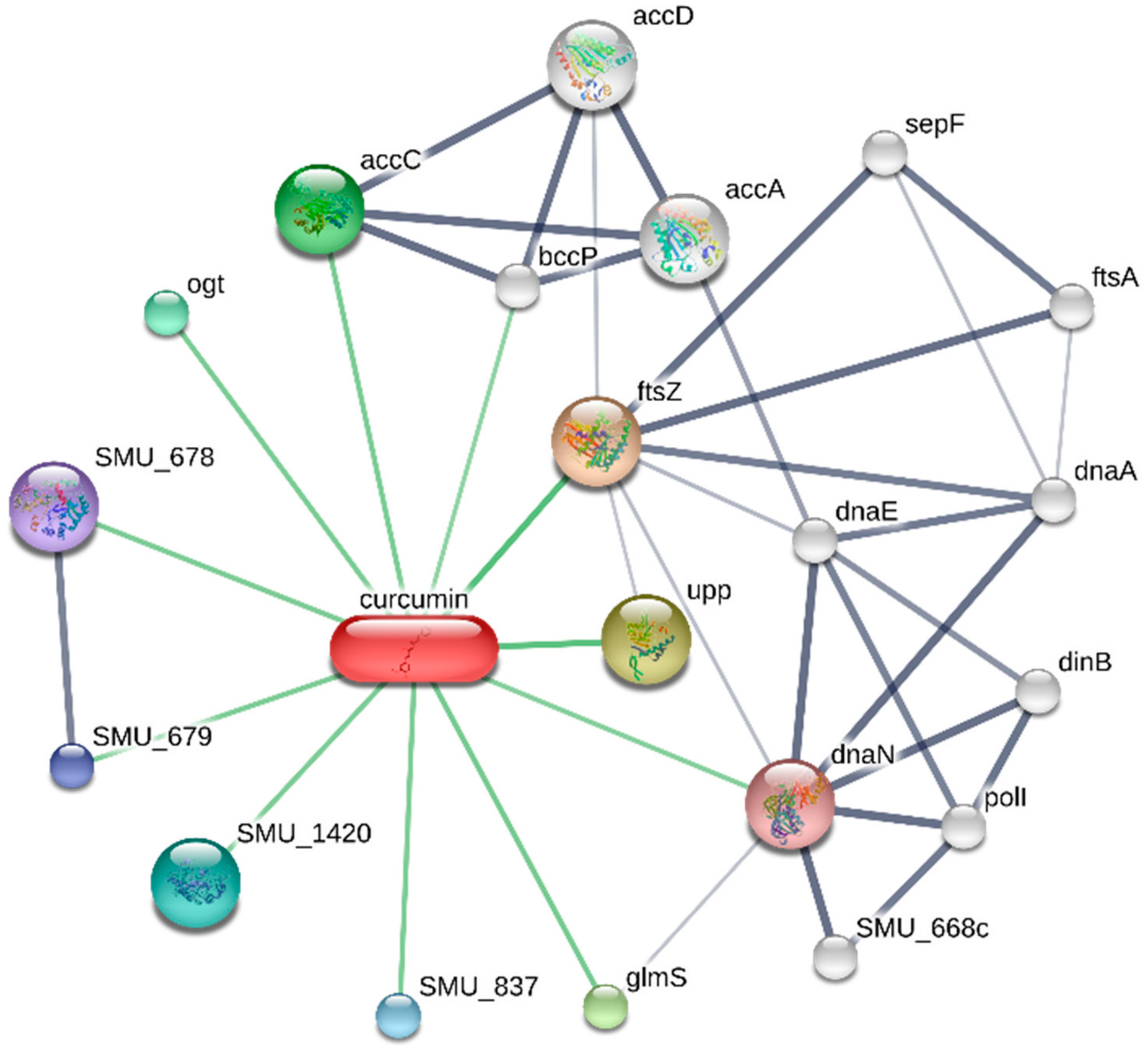
3.3. Target-Curcumin-Metabolic Pathways Network in Dental Caries
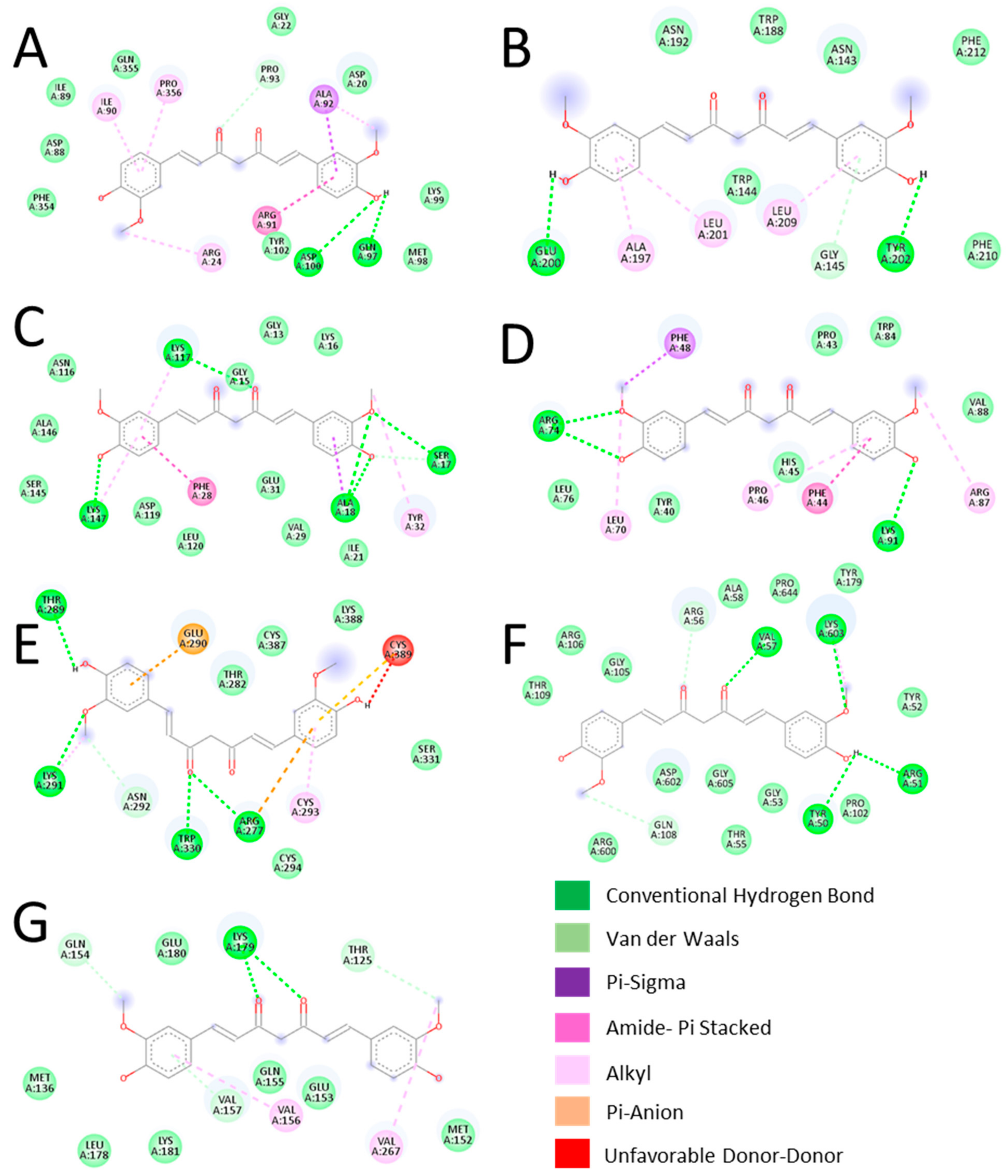
3.4. Validation of Key Proteins Using Molecular Docking
3.5. Effect of Curcumin on Streptococcus mutans
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Machiulskiene, V.; Campus, G.; Carvalho, J.C.; Dige, I.; Ekstrand, K.R.; Jablonski-Momeni, A.; Maltz, M.; Manton, D.J.; Martignon, S.; Martinez-Mier, E.A.; et al. Terminology of Dental Caries and Dental Caries Management: Consensus Report of a Workshop Organized by ORCA and Cariology Research Group of IADR. Caries Res. 2020, 54, 7–14. [Google Scholar] [CrossRef]
- Guzman-Flores, J.M.; Martínez-Esquivias, F.; Sarai Becerra-Ruiz, J.; Berenice Vázquez-Rodríguez, S. Integration of Proteomic Data Obtained from the Saliva of Children with Caries through Bioinformatic Analysis. Curr. Proteom. 2023, 20, 51–61. [Google Scholar] [CrossRef]
- Pitts, N.B.; Twetman, S.; Fisher, J.; Marsh, P.D. Understanding dental caries as a non-communicable disease. Br. Dent. J. 2021, 231, 749–753. [Google Scholar] [CrossRef]
- Mathur, V.P.; Dhillon, J.K. Dental Caries: A Disease Which Needs Attention. Indian J. Pediatr. 2018, 85, 202–206. [Google Scholar] [CrossRef]
- Milutinovici, R.A.; Chioran, D.; Buzatu, R.; Macasoi, I.; Razvan, S.; Chioibas, R.; Corlan, I.V.; Tanase, A.; Horia, C.; Popovici, R.A.; et al. Vegetal Compounds as Sources of Prophylactic and Therapeutic Agents in Dentistry. Plants 2021, 10, 2148. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Kim, B.S.; Keum, K.S.; Yu, H.H.; Kim, Y.H.; Chang, B.S.; Ra, J.Y.; Moon, H.D.; Seo, B.R.; Choi, N.Y.; et al. Essential oil of Curcuma longa inhibits Streptococcus mutans biofilm formation. J. Food Sci. 2011, 76, H226–H230. [Google Scholar] [CrossRef]
- Li, B.; Pan, T.; Lin, H.; Zhou, Y. The enhancing antibiofilm activity of curcumin on Streptococcus mutans strains from severe early childhood caries. BMC Microbiol. 2020, 20, 286. [Google Scholar] [CrossRef] [PubMed]
- Silva-Gaona, G.O.; Guzmán-Flores, M.J.; Hernández-Ortiz, M.; Vargas-Ortiz, K.; Ramírez-Emiliano, J.; Encarnación-Guevara, S.; Pérez-Vázquez, V. Curcumin Reverts the Protein Differential Expression in the Liver of the Diabetic Obese db/db Mice. Curr. Proteom. 2022, 19, 39–50. [Google Scholar] [CrossRef]
- Guzman-Flores, J.M.; Arevalo-Caro, C.M.; Martinez-Esquivias, F.; Isiordia-Espinoza, M.A.; Franco-de la Torre, L. Molecular mechanism of curcumin on periodontitis: A pharmacological network study. J. Oral Biosci. 2023, 65, 379–385. [Google Scholar] [CrossRef]
- Arellano-Rodriguez, N.C.; Alvarez-Quezada, O.A.; Benavides, P.Z.; Vargas-Alanis, G.; Franco-Molina, M.; Zamora-Avila, D.; Arellano-Rodriguez, M.; Saavedra-Alonso, S.; Izaguirre-Alvarez, J.M.; Rodriguez-Padilla, C. Curcumin Sensitizes 4T1 Murine Breast Cancer Cells to Cisplatin Through PAR4 Secretion. In Vivo 2022, 36, 2767–2773. [Google Scholar] [CrossRef]
- Ehteshami, A.; Shirban, F.; Gharibpour, F.; Bagherniya, M.; Sathyapalan, T.; Sahebkar, A. Does Curcumin Have an Anticaries Effect? A Systematic Review of In Vitro Studies. Adv. Exp. Med. Biol. 2021, 1291, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Nogales, C.; Mamdouh, Z.M.; List, M.; Kiel, C.; Casas, A.I.; Schmidt, H. Network pharmacology: Curing causal mechanisms instead of treating symptoms. Trends Pharmacol. Sci. 2022, 43, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Jiashuo, W.U.; Fangqing, Z.; Zhuangzhuang, L.I.; Weiyi, J.; Yue, S. Integration strategy of network pharmacology in Traditional Chinese Medicine: A narrative review. J. Tradit. Chin. Med. 2022, 42, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Wiegers, T.C.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2023. Nucleic Acids Res. 2023, 51, D1257–D1262. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- McLachlan, J.L.; Smith, A.J.; Bujalska, I.J.; Cooper, P.R. Gene expression profiling of pulpal tissue reveals the molecular complexity of dental caries. Biochim. Biophys. Acta 2005, 1741, 271–281. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Killcoyne, S.; Carter, G.W.; Smith, J.; Boyle, J. Cytoscape: A community-based framework for network modeling. Methods Mol. Biol. 2009, 563, 219–239. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef]
- Stanzione, F.; Giangreco, I.; Cole, J.C. Use of molecular docking computational tools in drug discovery. Prog. Med. Chem. 2021, 60, 273–343. [Google Scholar] [CrossRef] [PubMed]
- Loreto, C.; Psaila, A.; Musumeci, G.; Castorina, S.; Leonardi, R. Apoptosis activation in human carious dentin. An immunohistochemical study. Eur. J. Histochem. 2015, 59, 2513. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, N.; Whitford, G.M.; Bartlett, J.D.; Suzuki, M. Curcumin suppresses cell growth and attenuates fluoride-mediated Caspase-3 activation in ameloblast-like LS8 cells. Environ. Pollut. 2021, 273, 116495. [Google Scholar] [CrossRef] [PubMed]
- Bojanich, M.A.; Calderon, R.O. Streptococcus mutans membrane lipid composition: Virulence factors and structural parameters. Arch. Oral Biol. 2017, 81, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Casamada, C.; Molina-Frechero, N.; Espinosa-Cristobal, L.F.; Garcia-Lopez, S.; Castaneda-Castaneira, E. Polymorphisms associated with dental caries in pediatric populations: A systematic review. Rev. Med. Inst. Mex. Seguro Soc. 2023, 61, 502–508. [Google Scholar] [CrossRef]
- Gur, E.; Keles, S.; Cevik, O. Concentrations of interleukin-32, interleukin −10, interleukin −6, and TNF-alfa are higher in saliva of children with early childhood caries. Pediatr. Dent. J. 2023, 33, 116–124. [Google Scholar] [CrossRef]
- Khorasani, M.M.Y.; Andam-Shahsavari, P.; Zainodini, N.; Khoramdelazad, H.; Nosratabadi, R. Association of S100 calcium-binding protein A12, receptor for advanced glycation endproducts, and nuclear factor-κB expression with inflammation in pulp tissues from tooth caries. J. Investig. Clin. Dent. 2018, 9, e12290. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Flores, L.M.; Lopez-Briones, S.; Macias-Cervantes, M.H.; Ramirez-Emiliano, J.; Perez-Vazquez, V. A PPARgamma, NF-kappaB and AMPK-dependent mechanism may be involved in the beneficial effects of curcumin in the diabetic db/db mice liver. Molecules 2014, 19, 8289–8302. [Google Scholar] [CrossRef] [PubMed]
- Sahebkar, A.; Cicero, A.F.G.; Simental-Mendia, L.E.; Aggarwal, B.B.; Gupta, S.C. Curcumin downregulates human tumor necrosis factor-alpha levels: A systematic review and meta-analysis ofrandomized controlled trials. Pharmacol. Res. 2016, 107, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Cooper, P.R.; Friedlander, L.T.; Seo, B.; Rizwan, S.B.; Rich, A.M.; Hussaini, H.M. Potentiality and Inflammatory Marker Expression Are Maintained in Dental Pulp Cell Cultures from Carious Teeth. Int. J. Mol. Sci. 2022, 23, 9425. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Chen, Y.; Zhu, D.; Chen, G.; Chen, F.; Xu, N.; Barakat, K.; Zheng, J.; Xie, X.; Chen, R. Curcumin inhibits high glucose oxidative stress and apoptosis in pancreatic beta cells via CHOP/PCG-1a and pERK1/2. Front. Biosci. 2020, 25, 1974–1984. [Google Scholar] [CrossRef] [PubMed]
- Ke, D.; Xu, H.; Han, J.; Dai, H.; Wang, X.; Luo, J.; Yu, Y.; Xu, J. Curcumin suppresses RANKL-induced osteoclast precursor autophagy in osteoclastogenesis by inhibiting RANK signaling and downstream JNK-BCL2-Beclin1 pathway. Biomed. J. 2023, 47, 100605. [Google Scholar] [CrossRef] [PubMed]
- da SiLva, C.D.; da Silva, I.B.; Costa, V.; Carvalho, Y.R.; Kaminagakura, E. The relationship between ras protein and odontogenic lesions. Braz. Dent. Sci. 2018, 21, 307–314. [Google Scholar] [CrossRef]
- Wu, C.S.; Wu, S.Y.; Chen, H.C.; Chu, C.A.; Tang, H.H.; Liu, H.S.; Hong, Y.R.; Huang, C.F.; Huang, G.C.; Su, C.L. Curcumin functions as a MEK inhibitor to induce a synthetic lethal effect on KRAS mutant colorectal cancer cells receiving targeted drug regorafenib. J. Nutr. Biochem. 2019, 74, 108227. [Google Scholar] [CrossRef]
- Yan, G.; Huang, W.; Xue, H.; Jia, Y.; Yang, D. Relationship between dental caries and salivary proteome by electrospray ionization ion-trap tandem mass spectrometry in children aged 6 to 8 years. Hua Xi Kou Qiang Yi Xue Za Zhi Huaxi Kouqiang Yixue Zazhi West China J. Stomatol. 2014, 32, 297–302. [Google Scholar]
- Tong, W.; Wang, Q.; Sun, D.; Suo, J. Curcumin suppresses colon cancer cell invasion via AMPK-induced inhibition of NF-κB, uPA activator and MMP9. Oncol. Lett. 2016, 12, 4139–4146. [Google Scholar] [CrossRef]
- Gómez-García, A.P.; López-Vidal, Y.; Pinto-Cardoso, S.; Aguirre-García, M.M. Overexpression of proinflammatory cytokines in dental pulp tissue and distinct bacterial microbiota in carious teeth of Mexican Individuals. Front. Cell. Infect. Microbiol. 2022, 12, 958722. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.P.; Song, T.Z.; Zhu, Y.Y.; Wu, L.L.; Zhang, X.; Zhou, J.Y.; Li, Z.Q. Association of ENAM, TUFT1, MMP13, IL1B, IL10 and IL1RN gene polymorphism and dental caries susceptibility in Chinese children. J. Int. Med. Res. 2019, 47, 1696–1704. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.L.; Faustoferri, R.C.; Quivey, R.G. Acid-adaptive mechanisms of Streptococcus mutans—The more we know, the more we don’t. Mol. Oral Microbiol. 2017, 32, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Zeng, L.; Burne, R.A. Fueling the caries process: Carbohydrate metabolism and gene regulation by Streptococcus mutans . J. Oral Microbiol. 2014, 6, 24878. [Google Scholar] [CrossRef]
| Gene | Protein | Function |
|---|---|---|
| MAPK1 | Mitogen-activated protein kinase 1 | MAPK1 is involved in cell growth, adhesion, survival, and differentiation through the regulation of transcription, translation, and cytoskeletal rearrangements. |
| BCL2 | Apoptosis regulator Bcl-2 | Regulates cell death by controlling the mitochondrial membrane permeability. |
| KRAS | KRAS proto-oncogene, GTPase | It plays an important role in the regulation of cell proliferation |
| CXCL8 | Interleukin-8 | Chemotactic factor that mediates inflammatory response by attracting neutrophils, basophils, and T-cells to clear pathogens and protect the host from infection. |
| TGFB1 | Transforming growth factor beta-1 | Plays an important role in bone remodeling: acts as a potent stimulator of osteoblastic bone formation, causing chemotaxis, proliferation, and differentiation in committed osteoblasts. |
| MMP9 | Matrix metalloproteinase-9 | Could play a role in bone osteoclastic resorption. |
| IL1B | Interleukin-1 beta | It induces prostaglandin synthesis, neutrophil influx and activation, T-cell activation and cytokine production, B-cell activation and antibody production, and fibroblast proliferation and collagen production. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guzmán-Flores, J.M.; Pérez-Reyes, Á.; Vázquez-Jiménez, S.I.; Isiordia-Espinoza, M.A.; Martínez-Esquivias, F. A Docking and Network Pharmacology Study on the Molecular Mechanisms of Curcumin in Dental Caries and Streptococcus mutans. Dent. J. 2024, 12, 153. https://doi.org/10.3390/dj12060153
Guzmán-Flores JM, Pérez-Reyes Á, Vázquez-Jiménez SI, Isiordia-Espinoza MA, Martínez-Esquivias F. A Docking and Network Pharmacology Study on the Molecular Mechanisms of Curcumin in Dental Caries and Streptococcus mutans. Dentistry Journal. 2024; 12(6):153. https://doi.org/10.3390/dj12060153
Chicago/Turabian StyleGuzmán-Flores, Juan Manuel, Ángel Pérez-Reyes, Sonia Isela Vázquez-Jiménez, Mario Alberto Isiordia-Espinoza, and Fernando Martínez-Esquivias. 2024. "A Docking and Network Pharmacology Study on the Molecular Mechanisms of Curcumin in Dental Caries and Streptococcus mutans" Dentistry Journal 12, no. 6: 153. https://doi.org/10.3390/dj12060153
APA StyleGuzmán-Flores, J. M., Pérez-Reyes, Á., Vázquez-Jiménez, S. I., Isiordia-Espinoza, M. A., & Martínez-Esquivias, F. (2024). A Docking and Network Pharmacology Study on the Molecular Mechanisms of Curcumin in Dental Caries and Streptococcus mutans. Dentistry Journal, 12(6), 153. https://doi.org/10.3390/dj12060153









