Investigating Transcriptomic Induction of Resistance and/or Virulence in Listeria monocytogenes Cells Surviving Sublethal Antimicrobial Exposure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
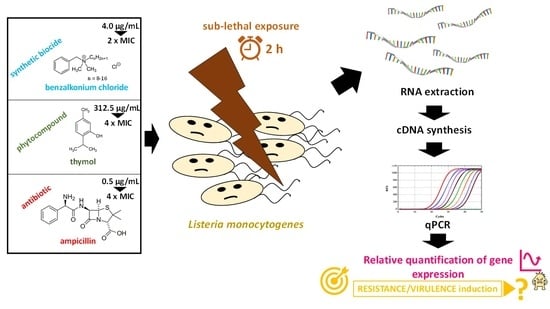
2.2. Chemical Antimicrobials (BAC, THY and AMP)
2.3. Determination of Minimum Inhibitory Concentration (MIC)
2.4. Sublethal Antimicrobial Exposure and RNA Extraction
2.5. Reverse Transcription (cDNA Synthesis)
2.6. qPCR for Quantitation of mRNA Transcripts
2.7. Statistical Analyses for Differential Gene Expression
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jordan, K.; McAuliffe, O. Listeria monocytogenes in foods. Adv. Food Nutr. Res. 2018, 86, 181–213. [Google Scholar] [CrossRef]
- EFSA; ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, 6406. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- NicAogáin, K.; O’Byrne, C.P. The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front. Microbiol. 2016, 7, 1865. [Google Scholar] [CrossRef] [Green Version]
- Soni, K.A.; Nannapaneni, R.; Tasara, T. The contribution of transcriptomic and proteomic analysis in elucidating stress adaptation responses of Listeria monocytogenes. Foodborne Pathog. Dis. 2011, 8, 843–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiensuu, T.; Guerreiro, D.N.; Oliveira, A.H.; O’Byrne, C.; Johansson, J. Flick of a switch: Regulatory mechanisms allowing Listeria monocytogenes to transition from a saprophyte to a killer. Microbiology 2019, 165, 819–833. [Google Scholar] [CrossRef] [PubMed]
- Dorey, A.; Marinho, C.; Piveteau, P.; O’Byrne, C. Role and regulation of the stress activated sigma factor sigma B (σB) in the saprophytic and host-associated life stages of Listeria monocytogenes. Adv. Appl. Microbiol. 2019, 106, 1–48. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, D.N.; Arcari, T.; O’Byrne, C.P. The σB-mediated general stress response of Listeria monocytogenes: Life and death decision making in a pathogen. Front. Microbiol. 2020, 11, 1505. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.L.; Ricke, S.C.; Donaldson, J.R. Establishment of Listeria monocytogenes in the gastrointestinal tract. Microorganisms 2019, 7, 75. [Google Scholar] [CrossRef] [Green Version]
- Gahan, C.G.; Hill, C. Listeria monocytogenes: Survival and adaptation in the gastrointestinal tract. Front. Cell. Infect. Microbiol. 2014, 4, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radoshevich, L.; Cossart, P. Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 2018, 16, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Rolhion, N.; Cossart, P. How the study of Listeria monocytogenes has led to new concepts in biology. Future Microbiol. 2017, 12, 621–638. [Google Scholar] [CrossRef]
- Bruno, J.C., Jr.; Freitag, N.E. Listeria monocytogenes adapts to long-term stationary phase survival without compromising bacterial virulence. FEMS Microbiol. Lett. 2011, 323, 171–179. [Google Scholar] [CrossRef] [Green Version]
- Gaballa, A.; Guariglia-Oropeza, V.; Wiedmann, M.; Boor, K.J. Cross talk between SigB and PrfA in Listeria monocytogenes facilitates transitions between extra- and intracellular environments. Microbiol. Mol. Biol. Rev. 2019, 83, e00034-19. [Google Scholar] [CrossRef]
- Matereke, L.T.; Okoh, A.I. Listeria monocytogenes virulence, antimicrobial resistance and environmental persistence: A review. Pathogens 2020, 9, 528. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Molfeta, C.; Panagiotopoulou, O.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Expression of Listeria monocytogenes key virulence genes during growth in liquid medium, on rocket and melon at 4, 10 and 30 °C. Food Microbiol. 2016, 55, 7–15. [Google Scholar] [CrossRef]
- Rieu, A.; Guzzo, J.; Piveteau, P. Sensitivity to acetic acid, ability to colonize abiotic surfaces and virulence potential of Listeria monocytogenes EGD-e after incubation on parsley leaves. J. Appl. Microbiol. 2010, 108, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Jespersen, L. Expression of virulence-related genes in Listeria monocytogenes grown on danish hard cheese as affected by NaCl Content. Foodborne Pathog. Dis. 2015, 12, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Mataragas, M.; Rovetto, F.; Bellio, A.; Alessandria, V.; Rantsiou, K.; Decastelli, L.; Cocolin, L. Differential gene expression profiling of Listeria monocytogenes in Cacciatore and Felino salami to reveal potential stress resistance biomarkers. Food Microbiol. 2015, 46, 408–417. [Google Scholar] [CrossRef]
- Olesen, I.; Thorsen, L.; Jespersen, L. Relative transcription of Listeria monocytogenes virulence genes in liver pâtés with varying NaCl content. Int. J. Food Microbiol. 2010, 141 (Suppl. 1), S60–S68. [Google Scholar] [CrossRef]
- Pérez-Baltar, A.; Alía, A.; Rodríguez, A.; Córdoba, J.J.; Medina, M.; Montiel, R. Impact of water activity on the inactivation and gene expression of Listeria monocytogenes during refrigerated storage of pressurized dry-cured ham. Foods 2020, 9, 1092. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Greppi, A.; Garosi, M.; Acquadro, A.; Mataragas, M.; Cocolin, L. Strain dependent expression of stress response and virulence genes of Listeria monocytogenes in meat juices as determined by microarray. Int. J. Food Microbiol. 2012, 152, 116–122. [Google Scholar] [CrossRef]
- Zolfaghari, M.; Rezaei, M.; Mohabbati Mobarez, A.; Forozandeh Moghaddam, M.; Hosseini, H.; Khezri, M. Virulence genes expression in viable but non-culturable state of Listeria monocytogenes in fish meat. Food Sci. Technol. Int. 2020, 26, 205–212. [Google Scholar] [CrossRef]
- Alía, A.; Rodríguez, A.; Andrade, M.J.; Gómez, F.M.; Córdoba, J.J. Combined effect of temperature, water activity and salt content on the growth and gene expression of Listeria monocytogenes in a dry-cured ham model system. Meat Sci. 2019, 155, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Makariti, I.P.; Printezi, A.; Kapetanakou, A.E.; Zeaki, N.; Skandamis, P.N. Investigating boundaries of survival, growth and expression of genes associated with stress and virulence of Listeria monocytogenes in response to acid and osmotic stress. Food Microbiol. 2015, 45, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Marras, L.; Carraro, V.; Sanna, C.; Sanna, A.; Ingianni, A.; Coroneo, V. Growth of Listeria monocytogenes in ready to eat salads at different storage temperatures and valuation of virulence genes expression. Ann. Ig. 2019, 31, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Olesen, I.; Vogensen, F.K.; Jespersen, L. Gene transcription and virulence potential of Listeria monocytogenes strains after exposure to acidic and NaCl stress. Foodborne Pathog. Dis. 2009, 6, 669–680. [Google Scholar] [CrossRef]
- Werbrouck, H.; Vermeulen, A.; Van Coillie, E.; Messens, W.; Herman, L.; Devlieghere, F.; Uyttendaele, M. Influence of acid stress on survival, expression of virulence genes and invasion capacity into Caco-2 cells of Listeria monocytogenes strains of different origins. Int. J. Food Microbiol. 2009, 134, 140–146. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Gkolfakis, P.; Patlaka, A.; Grounta, A.; Vourli, G.; Paramithiotis, S.; Touloumi, G.; Triantafyllou, K.; Drosinos, E.H. In vitro gene transcription of Listeria monocytogenes after exposure to human gastric and duodenal aspirates. J. Food Prot. 2020, 83, 89–100. [Google Scholar] [CrossRef]
- Jiang, L.; Olesen, I.; Andersen, T.; Fang, W.; Jespersen, L. Survival of Listeria monocytogenes in simulated gastrointestinal system and transcriptional profiling of stress- and adhesion-related genes. Foodborne Pathog. Dis. 2010, 7, 267–274. [Google Scholar] [CrossRef]
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to stress conditions encountered in food and food processing environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef] [Green Version]
- Gravesen, A.; Kallipolitis, B.; Holmstrøm, K.; Høiby, P.E.; Ramnath, M.; Knøchel, S. pbp2229-mediated nisin resistance mechanism in Listeria monocytogenes confers cross-protection to class IIa bacteriocins and affects virulence gene expression. Appl. Environ. Microbiol. 2004, 70, 1669–1679. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Jiang, X.; Zhang, Y.; Ji, S.; Gao, W.; Shi, L. Effect of benzalkonium chloride adaptation on sensitivity to antimicrobial agents and tolerance to environmental stresses in Listeria monocytogenes. Front. Microbiol. 2018, 9, 2906. [Google Scholar] [CrossRef] [Green Version]
- Giaouris, E.; Simões, M. Pathogenic biofilm formation in the food industry and alternative control strategies. In Handbook of Food Bioengineering, Foodborne Diseases; Holban, A.M., Grumezescu, A.M., Eds.; Academic Press (Elsevier): Amsterdam, The Netherlands, 2018; Volume 15. [Google Scholar] [CrossRef]
- Leistner, L. Basic aspects of food preservation by hurdle technology. Int. J. Food Microbiol. 2000, 55, 181–186. [Google Scholar] [CrossRef]
- Pupo, I.; Lepe, J.A.; Smani, Y.; Aznar, J. Comparison of the in vitro activity of ampicillin and moxifloxacin against Listeria monocytogenes at achievable concentrations in the central nervous system. J. Med. Microbiol. 2017, 66, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Suárez, J.V.; Ortiz, S.; López-Alonso, V. Potential impact of the resistance to quaternary ammonium disinfectants on the persistence of Listeria monocytogenes in food processing environments. Front Microbiol. 2016, 7, 638. [Google Scholar] [CrossRef] [Green Version]
- Kastbjerg, V.G.; Larsen, M.H.; Gram, L.; Ingmer, H. Influence of sublethal concentrations of common disinfectants on expression of virulence genes in Listeria monocytogenes. Appl. Environ. Microbiol. 2010, 76, 303–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, D.; Cerca, N.; Teixeira, P.; Oliveira, R.; Ceri, H.; Azeredo, J. Listeria monocytogenes and Salmonella enterica Enteritidis biofilms susceptibility to different disinfectants and stress-response and virulence gene expression of surviving cells. Microb. Drug Resist. 2011, 17, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Tamburro, M.; Ripabelli, G.; Vitullo, M.; Dallman, T.J.; Pontello, M.; Amar, C.F.; Sammarco, M.L. Gene expression in Listeria monocytogenes exposed to sublethal concentration of benzalkonium chloride. Comp. Immunol. Microbiol. Infect. Dis. 2015, 40, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.R.; Burall, L.S. Serotype to genotype: The changing landscape of listeriosis outbreak investigations. Food Microbiol. 2018, 75, 18–27. [Google Scholar] [CrossRef]
- Gerba, C.P. Quaternary ammonium biocides: Efficacy in application. Appl. Environ. Microbiol. 2015, 81, 464–469. [Google Scholar] [CrossRef] [Green Version]
- Kowalczyk, A.; Przychodna, M.; Sopata, S.; Bodalska, A.; Fecka, I. Thymol and thyme essential oil-new insights into selected therapeutic applications. Molecules 2020, 25, 4125. [Google Scholar] [CrossRef]
- Townsend, C.A. Convergent biosynthetic pathways to β-lactam antibiotics. Curr. Opin. Chem. Biol. 2016, 35, 97–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagliano, P.; Arslan, F.; Ascione, T. Epidemiology and treatment of the commonest form of listeriosis: Meningitis and bacteraemia. Infez. Med. 2017, 25, 210–216. [Google Scholar]
- Kostoglou, D.; Tsaklidou, P.; Iliadis, I.; Garoufallidou, N.; Skarmoutsou, G.; Koulouris, I.; Giaouris, E. Advanced killing potential of thymol against a time and temperature optimized attached Listeria monocytogenes population in lettuce broth. Biomolecules 2021, 11, 397. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Toledo-Arana, A.; Dussurget, O.; Nikitas, G.; Sesto, N.; Guet-Revillet, H.; Balestrino, D.; Loh, E.; Gripenland, J.; Tiensuu, T.; Vaitkevicius, K.; et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature 2009, 459, 950–956. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bickel, D.R. Degrees of differential gene expression: Detecting biologically significant expression differences and estimating their magnitudes. Bioinformatics 2004, 20, 682–688. [Google Scholar] [CrossRef] [Green Version]
- Cusimano, M.G.; Di Stefano, V.; La Giglia, M.; Di Marco Lo Presti, V.; Schillaci, D.; Pomilio, F.; Vitale, M. Control of growth and persistence of Listeria monocytogenes and β-lactam-resistant Escherichia coli by thymol in food processing settings. Molecules 2020, 25, 383. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Melcón, C.; Riesco-Peláez, F.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Susceptibility of Listeria monocytogenes planktonic cultures and biofilms to sodium hypochlorite and benzalkonium chloride. Food Microbiol. 2019, 82, 533–540. [Google Scholar] [CrossRef] [PubMed]
- López-Alonso, V.; Ortiz, S.; Corujo, A.; Martínez-Suárez, J.V. Analysis of benzalkonium chloride resistance and potential virulence of Listeria monocytogenes isolates obtained from different stages of a poultry production chain in Spain. J. Food Prot. 2020, 83, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Minarovičová, J.; Véghová, A.; Mikulášová, M.; Chovanová, R.; Šoltýs, K.; Drahovská, H.; Kaclíková, E. Benzalkonium chloride tolerance of Listeria monocytogenes strains isolated from a meat processing facility is related to presence of plasmid-borne bcrABC cassette. Antonie Van Leeuwenhoek 2018, 111, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Mullapudi, S.; Siletzky, R.M.; Kathariou, S. Heavy-metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey-processing plants. Appl. Environ. Microbiol. 2008, 74, 1464–1468. [Google Scholar] [CrossRef] [Green Version]
- Rakic-Martinez, M.; Drevets, D.A.; Dutta, V.; Katic, V.; Kathariou, S. Listeria monocytogenes strains selected on ciprofloxacin or the disinfectant benzalkonium chloride exhibit reduced susceptibility to ciprofloxacin, gentamicin, benzalkonium chloride, and other toxic compounds. Appl. Environ. Microbiol. 2011, 77, 8714–8721. [Google Scholar] [CrossRef] [Green Version]
- Capita, R.; Felices-Mercado, A.; García-Fernández, C.; Alonso-Calleja, C. Characterization of Listeria monocytogenes originating from the Spanish meat-processing chain. Foods 2019, 8, 542. [Google Scholar] [CrossRef] [Green Version]
- Maćkiw, E.; Stasiak, M.; Kowalska, J.; Kucharek, K.; Korsak, D.; Postupolski, J. Occurrence and characteristics of Listeria monocytogenes in ready-to-eat meat products in Poland. J. Food Prot. 2020, 83, 1002–1009. [Google Scholar] [CrossRef]
- Maćkiw, E.; Korsak, D.; Kowalska, J.; Felix, B.; Stasiak, M.; Kucharek, K.; Antoszewska, A.; Postupolski, J. Genetic diversity of Listeria monocytogenes isolated from ready-to-eat food products in retail in Poland. Int. J. Food Microbiol. 2021, 358, 109397. [Google Scholar] [CrossRef]
- Bouymajane, A.; Rhazi Filali, F.; Oulghazi, S.; Lafkih, N.; Ed-Dra, A.; Aboulkacem, A.; El Allaoui, A.; Ouhmidou, B.; Moumni, M. Occurrence, antimicrobial resistance, serotyping and virulence genes of Listeria monocytogenes isolated from foods. Heliyon 2021, 7, e06169. [Google Scholar] [CrossRef] [PubMed]
- Di Pasqua, R.; Mamone, G.; Ferranti, P.; Ercolini, D.; Mauriello, G. Changes in the proteome of Salmonella enterica serovar Thompson as stress adaptation to sublethal concentrations of thymol. Proteomics 2010, 10, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Dubois-Brissonnet, F.; Naïtali, M.; Mafu, A.A.; Briandet, R. Induction of fatty acid composition modifications and tolerance to biocides in Salmonella enterica serovar Typhimurium by plant-derived terpenes. Appl. Environ. Microbiol. 2011, 77, 906–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepe, J.A.; Rodríguez-Villodres, A.; Martín-Gutiérrez, G.; Luque, R.; Aznar, J. In vitro study of synergy of ampicillin with ceftriaxone against Listeria monocytogenes. Rev. Esp. Quimioter. 2019, 32, 465–468. [Google Scholar] [PubMed]
- Cho, Y.; Kim, H.; Beuchat, L.R.; Ryu, J.H. Synergistic activities of gaseous oregano and thyme thymol essential oils against Listeria monocytogenes on surfaces of a laboratory medium and radish sprouts. Food Microbiol. 2020, 86, 103357. [Google Scholar] [CrossRef] [PubMed]
- Finkel, S.E. Long-term survival during stationary phase: Evolution and the GASP phenotype. Nat. Rev. Microbiol. 2006, 4, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Johny, A.K.; Amalaradjou, M.A.; Ananda Baskaran, S.; Kim, K.S.; Venkitanarayanan, K. Plant-derived antimicrobials reduce Listeria monocytogenes virulence factors in vitro, and down-regulate expression of virulence genes. Int. J. Food Microbiol. 2012, 157, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Drolia, R.; Bhunia, A.K. Crossing the intestinal barrier via Listeria Adhesion Protein and Internalin A. Trends Microbiol. 2019, 27, 408–425. [Google Scholar] [CrossRef]
- Braschi, G.; Serrazanetti, D.I.; Siroli, L.; Patrignani, F.; De Angelis, M.; Lanciotti, R. Gene expression responses of Listeria monocytogenes Scott A exposed to sub-lethal concentrations of natural antimicrobials. Int. J. Food Microbiol. 2018, 286, 170–178. [Google Scholar] [CrossRef]
- Kaplan Zeevi, M.; Shafir, N.S.; Shaham, S.; Friedman, S.; Sigal, N.; Nir Paz, R.; Boneca, I.G.; Herskovits, A.A. Listeria monocytogenes multidrug resistance transporters and cyclic di-AMP, which contribute to type I interferon induction, play a role in cell wall stress. J. Bacteriol. 2013, 195, 5250–5261. [Google Scholar] [CrossRef]
- Bowman, J.P.; Lee Chang, K.J.; Pinfold, T.; Ross, T. Transcriptomic and phenotypic responses of Listeria monocytogenes strains possessing different growth efficiencies under acidic conditions. Appl. Environ. Microbiol. 2010, 76, 4836–4850. [Google Scholar] [CrossRef] [Green Version]
- Köhler, S.; Leimeister-Wächter, M.; Chakraborty, T.; Lottspeich, F.; Goebel, W. The gene coding for protein p60 of Listeria monocytogenes and its use as a specific probe for Listeria monocytogenes. Infect. Immun. 1990, 58, 1943–1950. [Google Scholar] [CrossRef] [Green Version]
- Ben Slama, R.; Miladi, H.; Chaieb, K.; Bakhrouf, A. Survival of Listeria monocytogenes cells and the effect of extended frozen storage (−20 °C) on the expression of its virulence gene. Appl. Biochem. Biotechnol. 2013, 170, 1174–1183. [Google Scholar] [CrossRef] [PubMed]
- Dramsi, S.; Kocks, C.; Forestier, C.; Cossart, P. Internalin-mediated invasion of epithelial cells by Listeria monocytogenes is regulated by the bacterial growth state, temperature and the pleiotropic activator prfA. Mol. Microbiol. 1993, 9, 931–941. [Google Scholar] [CrossRef]
- Gahan, C.G.; O’Mahony, J.; Hill, C. Characterization of the groESL operon in Listeria monocytogenes: Utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect. Immun. 2001, 69, 3924–3932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osborne, S.E.; Brumell, J.H. Listeriolysin O: From bazooka to Swiss army knife. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pöntinen, A.; Markkula, A.; Lindström, M.; Korkeala, H. Two-component-system histidine kinases involved in growth of Listeria monocytogenes EGD-e at low temperatures. Appl. Environ. Microbiol. 2015, 81, 3994–4004. [Google Scholar] [CrossRef] [Green Version]
- Cotter, P.D.; Guinane, C.M.; Hill, C. The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob. Agents Chemother. 2002, 46, 2784–2790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Yu, T.; Xu, Y.; Wang, H.; Korkeala, H.; Shi, L. MdrL, a major facilitator superfamily efflux pump of Listeria monocytogenes involved in tolerance to benzalkonium chloride. Appl. Microbiol. Biotechnol. 2019, 103, 1339–1350. [Google Scholar] [CrossRef]
- Browning, D.F.; Busby, S.J. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016, 14, 638–650. [Google Scholar] [CrossRef] [Green Version]
- Roberfroid, S.; Vanderleyden, J.; Steenackers, H. Gene expression variability in clonal populations: Causes and consequences. Crit. Rev. Microbiol. 2016, 42, 969–984. [Google Scholar] [CrossRef]
- Arguedas-Villa, C.; Stephan, R.; Tasara, T. Evaluation of cold growth and related gene transcription responses associated with Listeria monocytogenes strains of different origins. Food Microbiol. 2010, 27, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, L.D.; Vinther, J. RNA-Seq for bacterial gene expression. Curr. Protoc. Nucleic Acid Chem. 2018, 73, e55. [Google Scholar] [CrossRef] [PubMed]
 ) or AMP (0.5 μg/mL;
) or AMP (0.5 μg/mL;  ), in comparison to the untreated controls (no antimicrobial exposure). Each bar represents the mean ± standard errors (n = 9). The statistically significant differences in genes’ expressions relative to the controls appear as asterisks (*) above the bars, while ⌂ denote the statistically significant differences in genes’ expressions between the two strains (P < 0.05).
), in comparison to the untreated controls (no antimicrobial exposure). Each bar represents the mean ± standard errors (n = 9). The statistically significant differences in genes’ expressions relative to the controls appear as asterisks (*) above the bars, while ⌂ denote the statistically significant differences in genes’ expressions between the two strains (P < 0.05).
 ) or AMP (0.5 μg/mL;
) or AMP (0.5 μg/mL;  ), in comparison to the untreated controls (no antimicrobial exposure). Each bar represents the mean ± standard errors (n = 9). The statistically significant differences in genes’ expressions relative to the controls appear as asterisks (*) above the bars, while ⌂ denote the statistically significant differences in genes’ expressions between the two strains (P < 0.05).
), in comparison to the untreated controls (no antimicrobial exposure). Each bar represents the mean ± standard errors (n = 9). The statistically significant differences in genes’ expressions relative to the controls appear as asterisks (*) above the bars, while ⌂ denote the statistically significant differences in genes’ expressions between the two strains (P < 0.05).
| s/n | Gene | Locus Tag † | Product Name | Gene Size (bp) | Primer Sequence ‡ (5’ → 3’) | Amplicon Size (bp) | Amplification Efficiency (%) | R2 |
|---|---|---|---|---|---|---|---|---|
| 1 | groEL | lmo2068 | molecular chaperone GroEL | 1629 | F: AAGTCCAGCGTTATGTGCGA | 145 | 104.72 | 1.00 |
| R: CGTAGCTGGTGGTGGTACTG | ||||||||
| 2 | hly | lmo0202 | listeriolysin O precursor | 1590 | F: TGCCAGGTAACGCGAGAAAT | 135 | 93.96 | 1.00 |
| R: TGGTGCCCCAGATGGAGATA | ||||||||
| 3 | iap | lmo0582 | invasion associated secreted endopeptidase | 1449 | F: GCCAGAGCCGTGGATGTTAT | 178 | 113.63 | 0.99 |
| R: TTCTGGCGCACAATACGCTA | ||||||||
| 4 | inlA | lmo0433 | internalin A | 2403 | F: AAATCCTGTGGCACCACCAA | 137 | 95.78 | 1.00 |
| R: TTGTGCTGGCTGAATTCCCA | ||||||||
| 5 | inlB | lmo0434 | internalin B | 1893 | F: CGCGAAGCCAAAACACCAAT | 146 | 106.12 | 1.00 |
| R: TTGGCGCTGACATAACGAGT | ||||||||
| 6 | lisK | lmo1378 | two-component sensor histidine kinase | 1452 | F: GATGTGCGTGATTACGGGGA | 113 | 105.96 | 1.00 |
| R: CCGAGGCCATTACCACCTTT | ||||||||
| 7 | mdrD | lmo0872 | antibiotic resistance protein | 1167 | F: ATCGCCGATGTTTAGCGGAA | 113 | 108.26 | 0.98 |
| R: CATTCGCAAAATGCCCACCA | ||||||||
| 8 | mdrL | lmo2377 | multidrug transporter | 1212 | F: CCGTTGCTTGCGCTTTATGT | 117 | 94.03 | 0.97 |
| R: TCCCCATTTTCGCGTCATCA | ||||||||
| 9 | prfA | lmo0200 | listeriolysin positive regulatory protein | 714 | F: CTGAGCTATGTGCGATGCCA | 138 | 101.96 | 0.98 |
| R: AGCTTGGCTCTATTTGCGGT | ||||||||
| 10 | σB (sigB) | lmo0895 | RNA polymerase sigma factor SigB | 780 | F: CTTCAAAGCTCGCCGCAAAT | 182 | 105.40 | 1.00 |
| R: CCATCATCCGTACCACCAACA | ||||||||
| 11 | tuf | lmo2653 | elongation factor Tu | 1188 | F: CCAATGTTGTCGCCAGCTTC | 149 | 101.00 | 1.00 |
| R: GCAACTGGACGTGTTGAACG | ||||||||
| 12 | gap | lmo2459 | glyceraldehyde-3-phosphate dehydrogenase | 1011 | F: AGCTGCTTCCATAGCTGCATT | 114 | 95.88 | 0.96 |
| R: TTAGACGGAGCTGCTCAACG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokkoni, E.-A.; Andritsos, N.; Sakarikou, C.; Michailidou, S.; Argiriou, A.; Giaouris, E. Investigating Transcriptomic Induction of Resistance and/or Virulence in Listeria monocytogenes Cells Surviving Sublethal Antimicrobial Exposure. Foods 2021, 10, 2382. https://doi.org/10.3390/foods10102382
Kokkoni E-A, Andritsos N, Sakarikou C, Michailidou S, Argiriou A, Giaouris E. Investigating Transcriptomic Induction of Resistance and/or Virulence in Listeria monocytogenes Cells Surviving Sublethal Antimicrobial Exposure. Foods. 2021; 10(10):2382. https://doi.org/10.3390/foods10102382
Chicago/Turabian StyleKokkoni, Eleni-Anna, Nikolaos Andritsos, Christina Sakarikou, Sofia Michailidou, Anagnostis Argiriou, and Efstathios Giaouris. 2021. "Investigating Transcriptomic Induction of Resistance and/or Virulence in Listeria monocytogenes Cells Surviving Sublethal Antimicrobial Exposure" Foods 10, no. 10: 2382. https://doi.org/10.3390/foods10102382
APA StyleKokkoni, E.-A., Andritsos, N., Sakarikou, C., Michailidou, S., Argiriou, A., & Giaouris, E. (2021). Investigating Transcriptomic Induction of Resistance and/or Virulence in Listeria monocytogenes Cells Surviving Sublethal Antimicrobial Exposure. Foods, 10(10), 2382. https://doi.org/10.3390/foods10102382