A Colorimetric Strategy Based on Aptamer-Catalyzed Hairpin Assembly for the On-Site Detection of Salmonella typhimurium in Milk
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. S. typhimurium Culture
2.3. Preparation of AuNPs
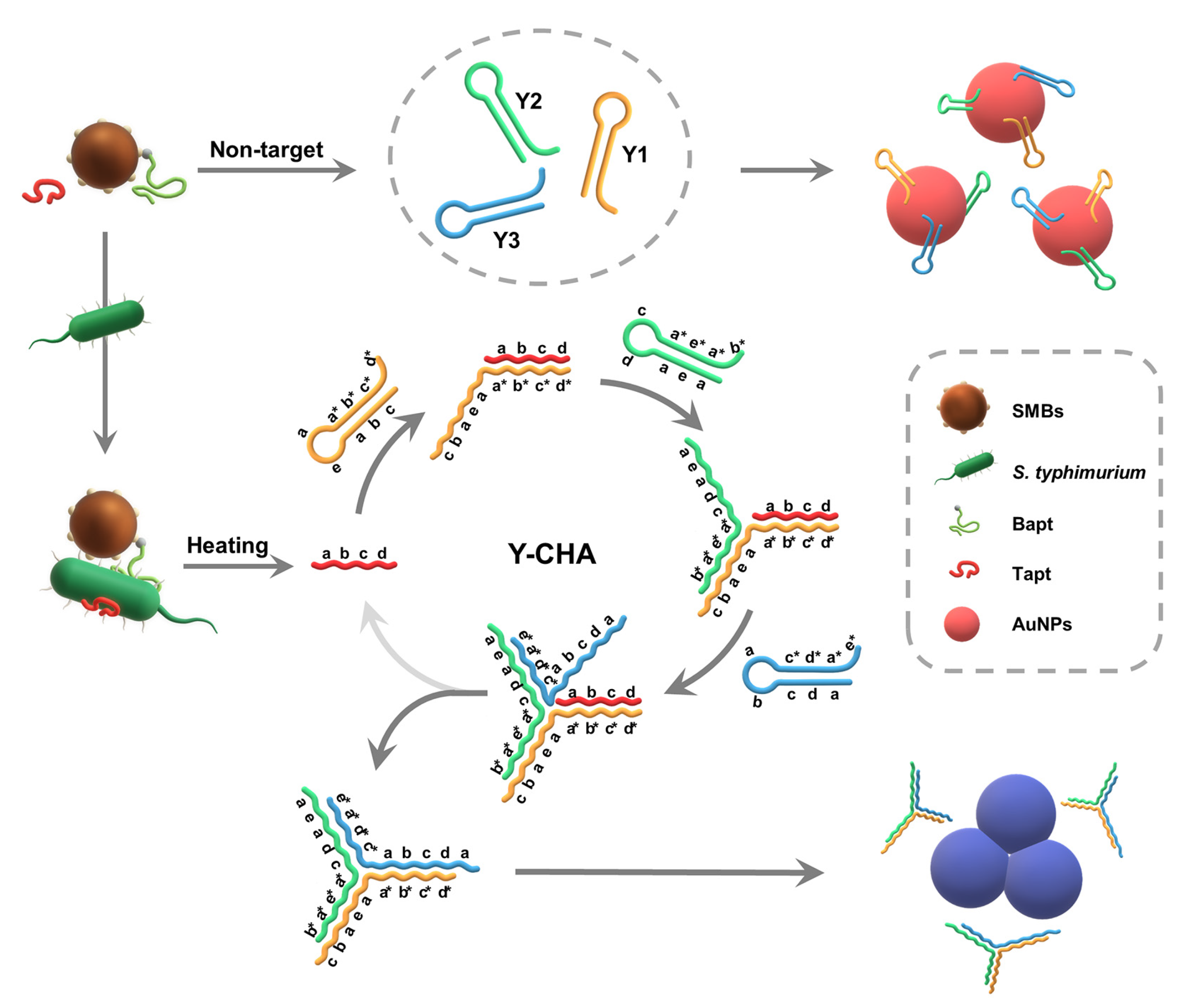
2.4. Y-CHA Circuit and Gel Electrophoresis
2.5. Synthesis of the Capture Complexes
2.6. Colorimetric Detection of S. typhimurium
2.7. Detection of S. typhimurium in Spiked Milk
3. Results and Discussion
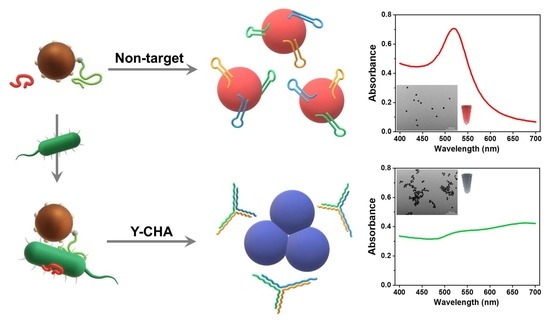
3.1. Principle of the Strategy
3.2. Gel Electrophoresis Analysis
3.3. Feasibility and Mechanism of AuNPs Colorimetric Detection
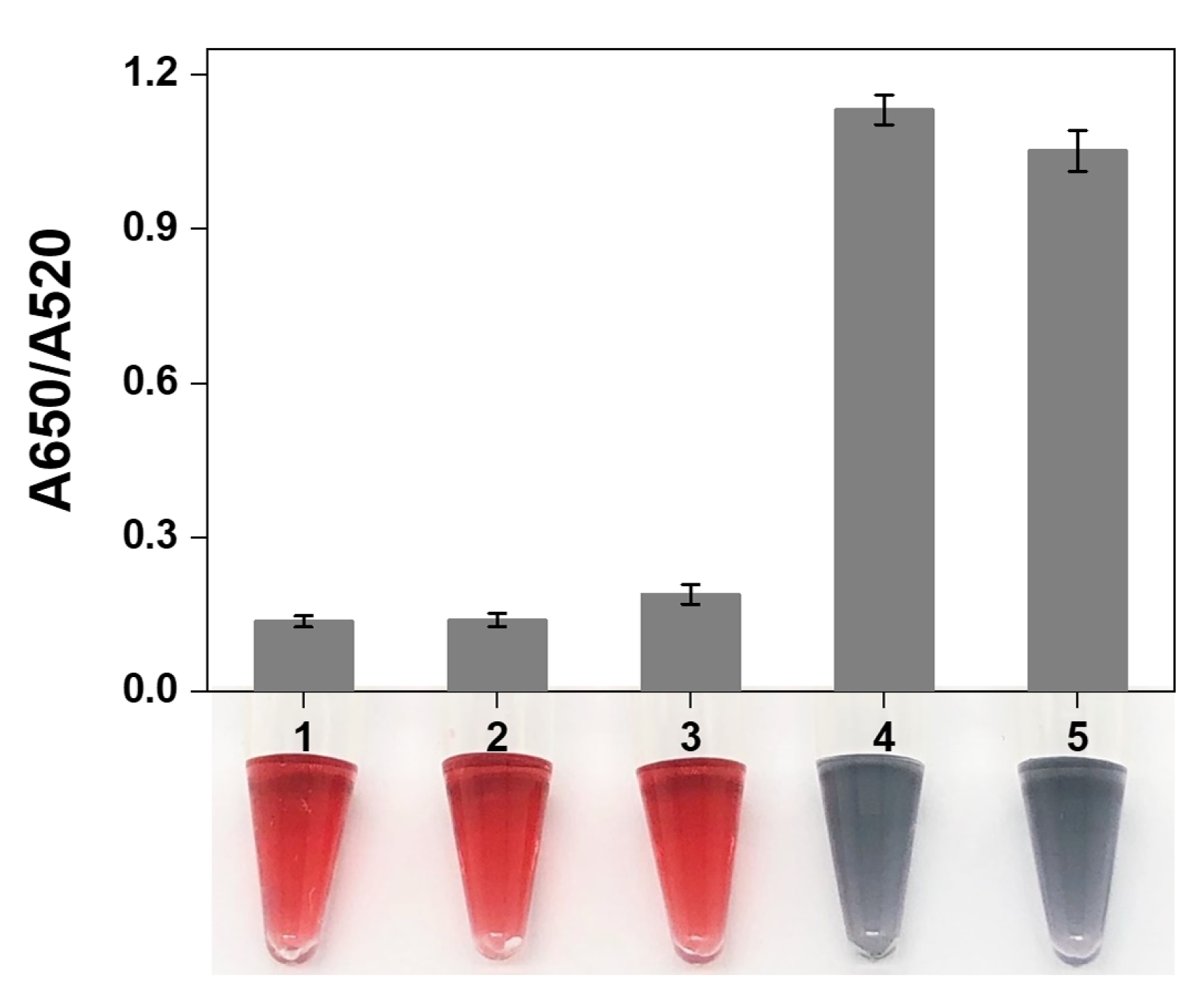
3.4. Finding the Optimum Conditions
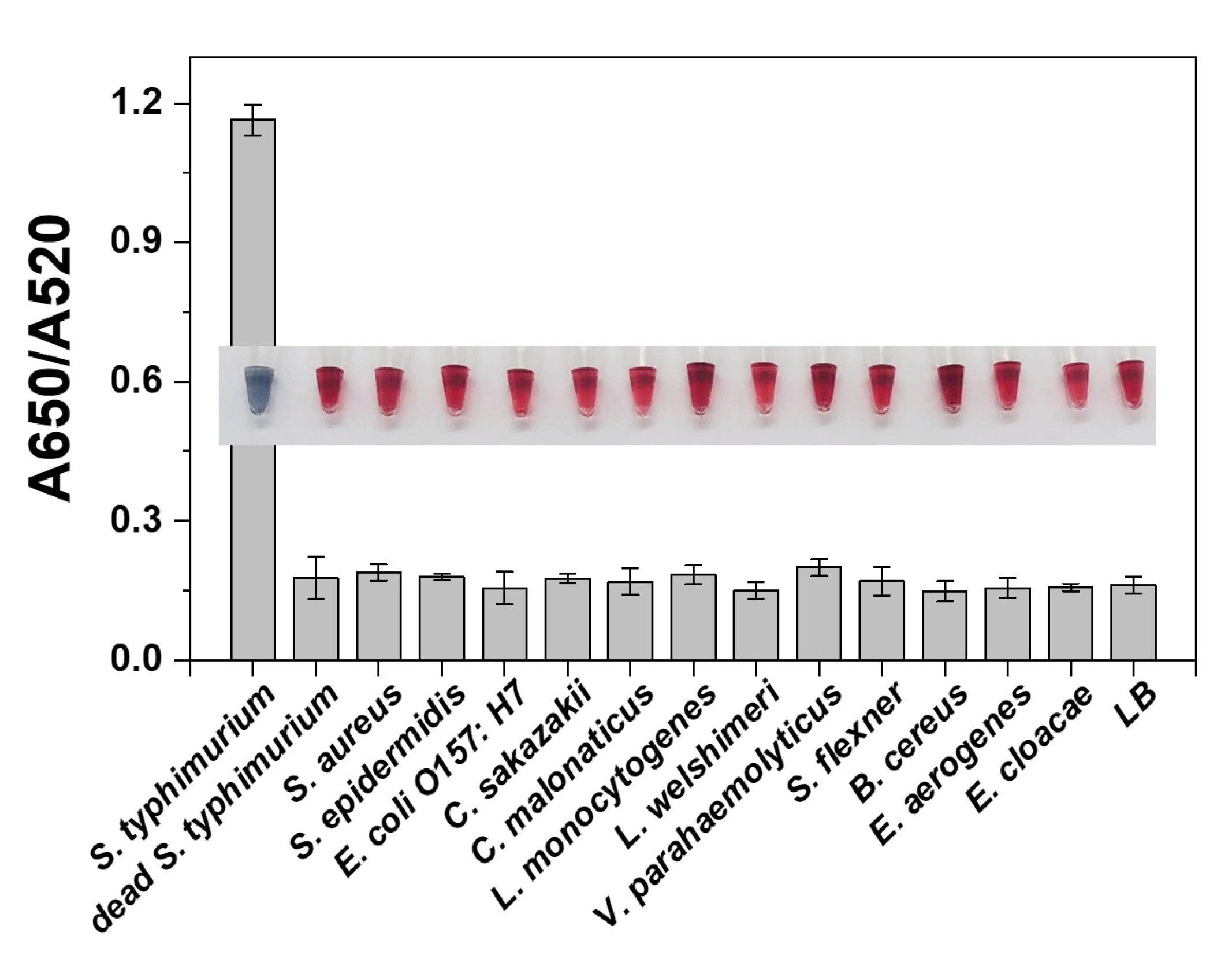
3.5. Analytical Performance of the Proposed Strategy
3.6. Authentic Sample Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne Illness Acquired in the United States-Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; World Health Organization: Genève, Switzerland, 2015. [Google Scholar]
- Batz, M.; Hoffmann, S.; Morris, J.G., Jr. Disease-Outcome Trees, EQ-5D Scores, and Estimated Annual Losses of Quality-Adjusted Life Years (QALYs) for 14 Foodborne Pathogens in the United States. Foodborne Pathog. Dis. 2014, 11, 395–402. [Google Scholar] [CrossRef]
- Gao, P.; Wang, L.; He, Y.; Wang, Y.; Yang, X.; Fu, S.; Qin, X.; Chen, Q.; Man, C.; Jiang, Y. An Enhanced Lateral Flow Assay Based on Aptamer–Magnetic Separation and Multifold AuNPs for Ultrasensitive Detection of Salmonella Typhimurium in Milk. Foods 2021, 10, 1605. [Google Scholar] [CrossRef] [PubMed]
- Robinson, E.; Travanut, M.; Fabre, L.; Larreche, S.; Ramelli, L.; Pascal, L.; Guinard, A.; Vincent, N.; Calba, C.; Meurice, L.; et al. Outbreak of Salmonella Newport associated with internationally distributed raw goats’ milk cheese, France, 2018. Epidemiol. Infect. 2020, 148, e180. [Google Scholar] [CrossRef]
- Fang, S.; Song, D.; Zhuo, Y.; Chen, Y.; Zhu, A.; Long, F. Simultaneous and sensitive determination of Escherichia coli O157:H7 and Salmonella Typhimurium using evanescent wave dual-color fluorescence aptasensor based on micro/nano size effect. Biosens. Bioelectron. 2021, 185, 113288. [Google Scholar] [CrossRef]
- Ren, Y.; Wei, J.; He, Y.; Wang, Y.; Bai, M.; Zhang, C.; Luo, L.; Wang, J.; Wang, Y. Ultrasensitive label-free immunochromatographic strip sensor for Salmonella determination based on salt-induced aggregated gold nanoparticles. Food Chem. 2021, 343, 128518. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, L.; Pang, L.; Fu, S.; Qin, X.; Chen, Q.; Man, C.; Jiang, Y. A novel fluorescent platform of DNA-stabilized silver nanoclusters based on exonuclease III amplification-assisted detection of Salmonella Typhimurium. Anal. Chim. Acta 2021, 1181, 338903. [Google Scholar] [CrossRef]
- Ren, Y.; Gao, P.; Song, Y.; Yang, X.; Yang, T.; Chen, S.; Fu, S.; Qin, X.; Shao, M.; Man, C.; et al. An aptamer-exonuclease III (Exo III)–assisted amplification-based lateral flow assay for sensitive detection of Escherichia coli O157:H7 in milk. J. Dairy Sci. 2021, 104, 8517–8529. [Google Scholar] [CrossRef]
- Jiang, Y.J.; Chen, S.H.; Zhao, Y.M.; Yang, X.Y.; Fu, S.Q.; McKillip, J.L.; Fox, E.M.; Man, C.X. Multiplex loop-mediated isothermal amplification-based lateral flow dipstick for simultaneous detection of 3 food-borne pathogens in powdered infant formula. J. Dairy Sci. 2020, 103, 4002–4012. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Choi, H.M.T.; Calvert, C.R.; Pierce, N.A. Programming biomolecular self-assembly pathways. Nature 2008, 451, 318–322. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhang, Y.; Zhao, Q.; Bo, S.; Zhao, J.; Luo, S.; Li, B.; Yan, X.; Vadgama, P.; Su, L.; et al. Bifunctional aptamer-mediated catalytic hairpin assembly for the sensitive and homogenous detection of rare cancer cells. Anal. Chim. Acta 2018, 1029, 58–64. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Y.; Xie, H.; Zhao, L.; Zheng, L.; Ye, H. Applications of Catalytic Hairpin Assembly Reaction in Biosensing. Small 2019, 15, 1902989. [Google Scholar] [CrossRef]
- Jung, C.; Ellington, A.D. Diagnostic Applications Applications of Nucleic Acid Circuits. Acc. Chem. Res. 2014, 47, 1825–1835. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Ellington, A.D.; Chen, X. Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods. Nucleic Acids Res. 2011, 39, e110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.; Song, Y.; Jang, K.; Choi, C.-H.; Na, S. Target switching catalytic hairpin assembly and gold nanoparticle colorimetric for EGFR mutant detection. Sens. Actuators B Chem. 2018, 261, 497–504. [Google Scholar] [CrossRef]
- He, H.; Dai, J.; Duan, Z.; Meng, Y.; Zhou, C.; Long, Y.; Zheng, B.; Du, J.; Guo, Y.; Xiao, D. Target-catalyzed autonomous assembly of dendrimer-like DNA nanostructures for enzyme-free and signal amplified colorimetric nucleic acids detection. Biosens. Bioelectron. 2016, 86, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Wu, Y.; Xu, S.; Tang, Y.; Zhang, X.; Wu, P. Improving the Signal-to-Background Ratio during Catalytic Hairpin Assembly through Both-End-Blocked DNAzyme. ACS Sens. 2018, 3, 1190–1195. [Google Scholar] [CrossRef]
- Zou, L.; Li, X.; Lai, Y. Colorimetric aptasensor for sensitive detection of kanamycin based on target-triggered catalytic hairpin assembly amplification and DNA-gold nanoparticle probes. Microchem. J. 2021, 162, 105858. [Google Scholar] [CrossRef]
- Ravan, H.; Norouzi, A.; Sanadgol, N.; Hosseinzadeh, E. Colorimetric nanoplatform for visual determination of cancer cells via target-catalyzed hairpin assembly actuated aggregation of gold nanoparticles. Microchim. Acta 2020, 187, 8. [Google Scholar] [CrossRef]
- Yu, L.; Song, Z.; Peng, J.; Yang, M.; Zhi, H.; He, H. Progress of gold nanomaterials for colorimetric sensing based on different strategies. Trac-Trends Anal. Chem. 2020, 127. [Google Scholar] [CrossRef]
- Xie, Y.; Huang, Y.; Li, J.; Wu, J. A trigger-based aggregation of aptamer-functionalized gold nanoparticles for colorimetry: An example on detection of Escherichia coli O157:H7. Sens. Actuators B Chem. 2021, 339, 129865. [Google Scholar] [CrossRef]
- Gupta, R.; Kumar, A.; Kumar, S.; Pinnaka, A.K.; Singhal, N.K. Naked eye colorimetric detection of Escherichia coli using aptamer conjugated graphene oxide enclosed Gold nanoparticles. Sens. Actuators B Chem. 2021, 329, 129100. [Google Scholar] [CrossRef]
- Yun, W.; Jiang, J.; Cai, D.; Zhao, P.; Liao, J.; Sang, G. Ultrasensitive visual detection of DNA with tunable dynamic range by using unmodified gold nanoparticles and target catalyzed hairpin assembly amplification. Biosens. Bioelectron. 2016, 77, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Yang, X.; Sun, S.; Wang, Q.; Wang, K.; Huang, J.; Liu, J.; He, L. Enzyme-Free Colorimetric Detection of DNA by Using Gold Nanoparticles and Hybridization Chain Reaction Amplification. Anal. Chem. 2013, 85, 7689–7695. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Ying, Y.; Ping, J. Colorimetric aggregation assay for kanamycin using gold nanoparticles modified with hairpin DNA probes and hybridization chain reaction-assisted amplification. Microchim. Acta 2019, 186, 448. [Google Scholar] [CrossRef]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and Design of Nucleic Acid Systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef]
- Joshi, R.; Janagama, H.; Dwivedi, H.P.; Kumar, T.M.A.S.; Jaykus, L.-A.; Schefers, J.; Sreevatsan, S. Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars. Mol. Cell. Probes 2009, 23, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat. Protoc. 2006, 1, 246–252. [Google Scholar] [CrossRef]
- Chen, S.H.; Yang, X.Y.; Fu, S.Q.; Qin, X.; Yang, T.; Man, C.X.; Jiang, Y.J. A novel AuNPs colorimetric sensor for sensitively detecting viable Salmonella typhimurium based on dual aptamers. Food Control 2020, 115, 8. [Google Scholar] [CrossRef]
- Tan, Z.J.; Chen, S.J. Nucleic acid helix stability: Effects of salt concentration, cation valence and size, and chain length. Biophys. J. 2006, 90, 1175–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Jin, J.; Du, Y.; Zhou, X.; Liang, H.; Jiang, W. Enhancing the stability of single-stranded DNA on gold nanoparticles as molecular machines through salt and acid regulation. J. Mater. Chem. B 2019, 7, 5554–5562. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Fang, X.; Liu, Y.; Kong, J.; Chen, Q. A hybridization chain reaction coupled with gold nanoparticles for allergen gene detection in peanut, soybean and sesame DNAs. Analyst 2019, 144, 3886–3891. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, H.; Li, X.; Ma, S.; Men, S.; Wei, H.; Cui, J.; Wang, H. A label-free fluorescent direct detection of live Salmonella typhimurium using cascade triple trigger sequences-regenerated strand displacement amplification and hairpin template-generated-scaffolded silver nanoclusters. Biosens. Bioelectron. 2017, 87, 1044–1049. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Zong, X.; Zheng, J.; Zhang, J.; Zhou, M.; Chen, Q.; Man, C.; Jiang, Y. A Colorimetric Strategy Based on Aptamer-Catalyzed Hairpin Assembly for the On-Site Detection of Salmonella typhimurium in Milk. Foods 2021, 10, 2539. https://doi.org/10.3390/foods10112539
Chen S, Zong X, Zheng J, Zhang J, Zhou M, Chen Q, Man C, Jiang Y. A Colorimetric Strategy Based on Aptamer-Catalyzed Hairpin Assembly for the On-Site Detection of Salmonella typhimurium in Milk. Foods. 2021; 10(11):2539. https://doi.org/10.3390/foods10112539
Chicago/Turabian StyleChen, Sihan, Xinran Zong, Jiapeng Zheng, Jiaxin Zhang, Mengyao Zhou, Qing Chen, Chaoxin Man, and Yujun Jiang. 2021. "A Colorimetric Strategy Based on Aptamer-Catalyzed Hairpin Assembly for the On-Site Detection of Salmonella typhimurium in Milk" Foods 10, no. 11: 2539. https://doi.org/10.3390/foods10112539
APA StyleChen, S., Zong, X., Zheng, J., Zhang, J., Zhou, M., Chen, Q., Man, C., & Jiang, Y. (2021). A Colorimetric Strategy Based on Aptamer-Catalyzed Hairpin Assembly for the On-Site Detection of Salmonella typhimurium in Milk. Foods, 10(11), 2539. https://doi.org/10.3390/foods10112539