Real-Time PCR Assay for the Detection and Quantification of Roe Deer to Detect Food Adulteration—Interlaboratory Validation Involving Laboratories in Austria, Germany, and Switzerland
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participating Laboratories
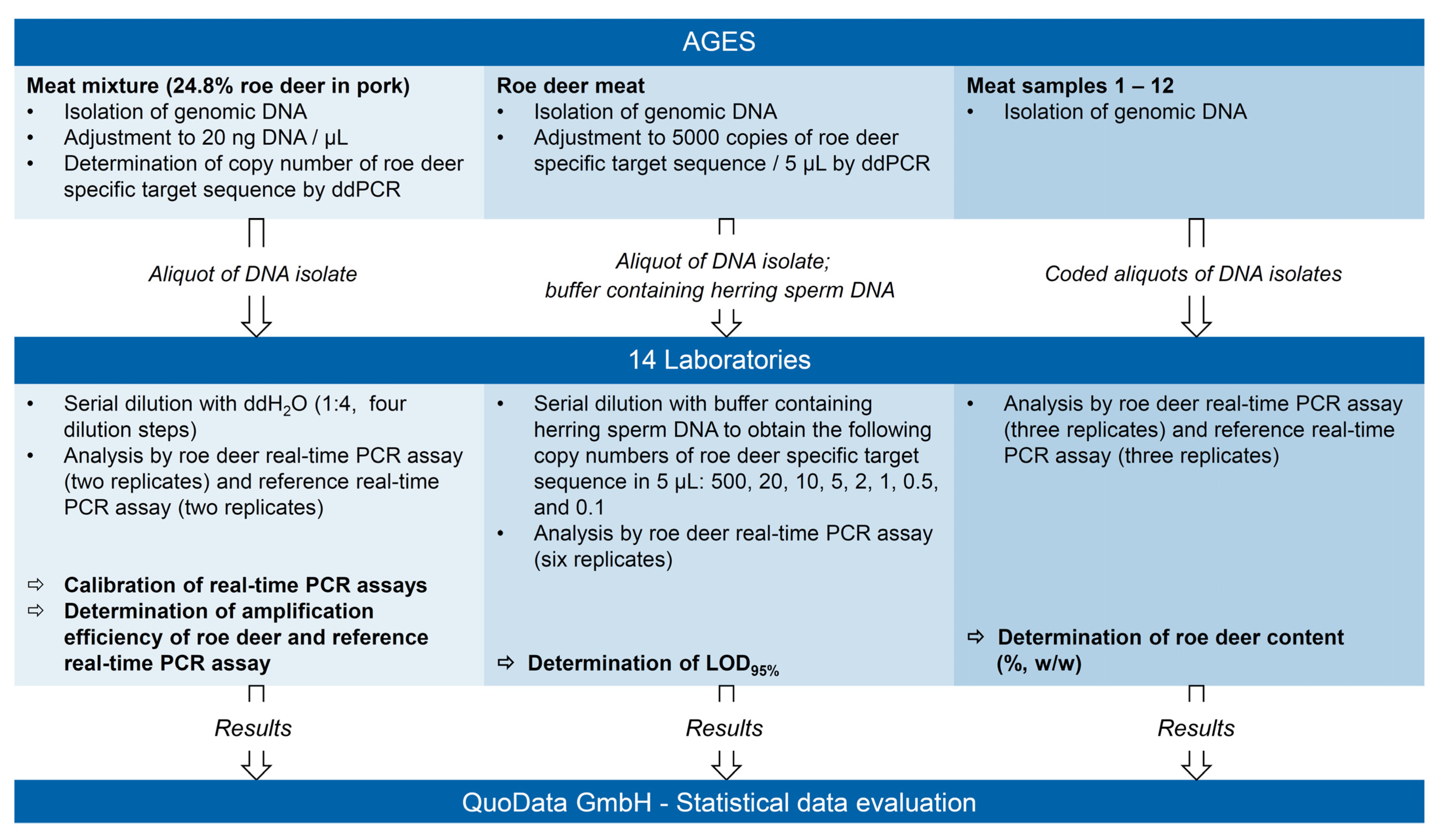
2.2. Design of the Interlaboratory Ring Trial
2.3. Meat Samples
2.4. Isolation of Genomic DNA
2.5. Real-Time PCR
2.6. Data Evaluation and Statistical Analysis
- Ct: Ct value
- d: intercept of the standard curve
- slope: slope of the standard curve
3. Results and Discussion
3.1. Amplification Efficiency
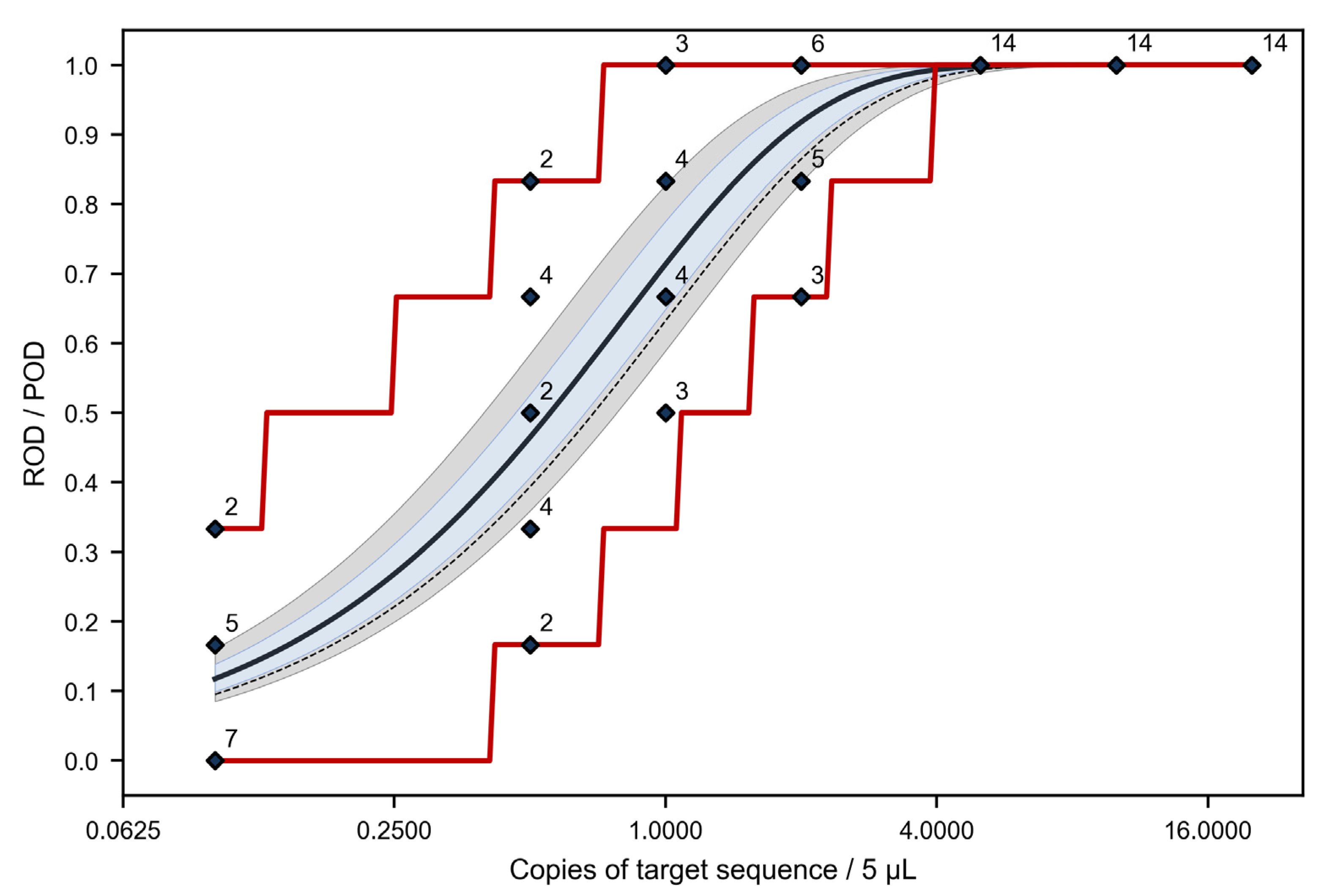
3.2. Level of Detection (LOD95%)
3.2.1. LOD95% According to Simplified Calculation Approaches
3.2.2. LOD95% Derived from the Mixed Model for the POD Curve
3.3. Analysis of Meat Samples
3.3.1. False Positive and False Negative Results
3.3.2. Quantitative Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoffman, L.C.; Wiklund, E. Game and venison—Meat for the modern consumer. Meat Sci. 2006, 74, 197–208. [Google Scholar] [CrossRef]
- Ballin, N.Z. Authentication of meat and meat products. Meat Sci. 2010, 86, 577–587. [Google Scholar] [CrossRef]
- Premanandh, J. Horse meat scandal—A wake-up call for regulatory authorities. Food Control 2013, 34, 568–569. [Google Scholar] [CrossRef]
- D’Amato, M.E.; Alechine, E.; Cloete, K.W.; Davison, S.; Corach, D. Where is the game? Wild meat products authentication in South Africa: A case study. Investig. Genet. 2013, 4, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatis, C.; Sarri, C.A.; Moutou, K.A.; Argyrakoulis, N.; Galara, I.; Godosopoulos, V.; Kolovos, M.; Liakou, C.; Stasinou, V.; Mamuris, Z. What do we think we eat? Single tracing method across foodstuff of animal origin found in Greek market. Food Res. Int. 2015, 69, 151–155. [Google Scholar] [CrossRef]
- Fajardo, V.; González, I.; López-Calleja, I.; Martín, I.; Rojas, M.; Hernández, P.E.; García, T.; Martín, R. Identification of meats from red deer (Cervus elaphus), fallow deer (Dama dama), and roe deer (Capreolus capreolus) using polymerase chain reaction targeting specific sequences from the mitochondrial 12S rRNA gene. Meat Sci. 2007, 76, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, V.; González, I.; Martín, I.; Rojas, M.; Hernández, P.E.; García, T.; Martín, R. Differentiation of European wild boar (Sus scrofa scrofa) and domestic swine (Sus scrofa domestica) meats by PCR analysis targeting the mitochondrial D-loop and the nuclear melanocortin receptor 1 (MC1R) genes. Meat Sci. 2008, 78, 314–322. [Google Scholar] [CrossRef]
- Rojas, M.; González, I.; Pavón, M.Á.; Pegels, N.; Lago, A.; Hernández, P.E.; García, T.; Martín, R. Novel TaqMan real-time polymerase chain reaction assay for verifying the authenticity of meat and commercial meat products from game birds. Food Addit. Contam. 2010, 27, 749–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amaral, J.S.; Santos, C.G.; Melo, V.S.; Oliveira, M.B.P.P.; Mafra, I. Authentication of a traditional game meat sausage (Alheira) by species-specific PCR assays to detect hare, rabbit, red deer, pork and cow meats. Food Res. Int. 2014, 60, 140–145. [Google Scholar] [CrossRef] [Green Version]
- Rak, L.; Knapik, K.; Bania, J.; Sujkowski, J.; Gadzinowski, A. Detection of roe deer, red deer, and hare meat in raw materials and processed products available in Poland. Eur. Food Res. Technol. 2014, 239, 189–194. [Google Scholar] [CrossRef]
- Codex Alimentarius Austriacus. Österreichisches Lebensmittelbuch, Codexkapitel/B14/Fleisch und Fleischerzeugnisse; Springer: Vienna, Austria, 2005. [Google Scholar]
- Druml, B.; Grandits, S.; Mayer, W.; Hochegger, R.; Cichna-Markl, M. Authenticity control of game meat products—A single method to detect and quantify adulteration of fallow deer (Dama dama), red deer (Cervus elaphus) and sika deer (Cervus nippon) by real-time PCR. Food Chem. 2015, 170, 508–517. [Google Scholar] [CrossRef]
- Druml, B.; Hochegger, R.; Cichna-Markl, M. Duplex real-time PCR assay for the simultaneous determination of the roe deer (Capreolus capreolus) and deer (sum of fallow deer, red deer and sika deer) content in game meat products. Food Control 2015, 57, 370–376. [Google Scholar] [CrossRef]
- Druml, B.; Mayer, W.; Cichna-Markl, M.; Hochegger, R. Development and validation of a TaqMan real-time PCR assay for the identification and quantification of roe deer (Capreolus capreolus) in food to detect food adulteration. Food Chem. 2015, 178, 319–326. [Google Scholar] [CrossRef]
- Kaltenbrunner, M.; Hochegger, R.; Cichna-Markl, M. Red deer (Cervus elaphus)-specific real-time PCR assay for the detection of food adulteration. Food Control 2018, 89, 157–166. [Google Scholar] [CrossRef]
- Kaltenbrunner, M.; Hochegger, R.; Cichna-Markl, M. Sika deer (Cervus nippon)-specific real-time PCR method to detect fraudulent labelling of meat and meat products. Sci. Rep. 2018, 8, 7236. [Google Scholar] [CrossRef] [PubMed]
- Kaltenbrunner, M.; Hochegger, R.; Cichna-Markl, M. Tetraplex real-time PCR assay for the simultaneous identification and quantification of roe deer, red deer, fallow deer and sika deer for deer meat authentication. Food Chem. 2018, 269, 486–494. [Google Scholar] [CrossRef]
- Kaltenbrunner, M.; Hochegger, R.; Cichna-Markl, M. Development and validation of a fallow deer (Dama dama)-specific TaqMan real-time PCR assay for the detection of food adulteration. Food Chem. 2018, 243, 82–90. [Google Scholar] [CrossRef]
- Kaltenbrunner, M.; Mayer, W.; Kerkhoff, K.; Epp, R.; Rüggeberg, H.; Hochegger, R.; Cichna-Markl, M. Differentiation between wild boar and domestic pig in food by targeting two gene loci by real-time PCR. Sci. Rep. 2019, 9, 9221. [Google Scholar] [CrossRef]
- Kaltenbrunner, M.; Mayer, W.; Kerkhoff, K.; Epp, R.; Rüggeberg, H.; Hochegger, R.; Cichna-Markl, M. Applicability of a duplex and four singleplex real-time PCR assays for the qualitative and quantitative determination of wild boar and domestic pig meat in processed food products. Sci. Rep. 2020, 10, 17243. [Google Scholar] [CrossRef]
- Ballin, N.Z.; Vogensen, F.K.; Karlsson, A.H. Species determination—Can we detect and quantify meat adulteration? Meat Sci. 2009, 83, 165–174. [Google Scholar] [CrossRef]
- Laube, I.; Zagon, J.; Spiegelberg, A.; Butschke, A.; Kroh, L.W.; Broll, H. Development and design of a ‘ready-to-use’ reaction plate for a PCR-based simultaneous detection of animal species used in foods. Int. J. Food Sci. Tech. 2007, 42, 9–17. [Google Scholar] [CrossRef]
- Eugster, A.; Ruf, J.; Rentsch, J.; Hübner, P.; Köppel, R. Quantification of beef and pork fraction in sausages by real-time PCR analysis: Results of an interlaboratory trial. Eur. Food Res. Technol. 2008, 227, 17–20. [Google Scholar] [CrossRef]
- Eugster, A.; Ruf, J.; Rentsch, J.; Köppel, R. Quantification of beef, pork, chicken and turkey proportions in sausages: Use of matrix-adapted standards and comparison of single versus multiplex PCR in an interlaboratory trial. Eur. Food Res. Technol. 2009, 230, 55–61. [Google Scholar] [CrossRef]
- Taverniers, I.; De Loose, M.; Van Bockstaele, E. Trends in quality in the analytical laboratory. II. Analytical method validation and quality assurance. Trends Anal. Chem. 2004, 23, 535–552. [Google Scholar] [CrossRef]
- ISO 21571. Foodstuffs – Methods of Analysis for the Detection of Genetically Modified Organisms and Derived Products: Nucleic Acid Extraction. Annex A3; International Organization for Standardization: Geneva, Switzerland, 2005. [Google Scholar]
- Uhlig, S.; Frost, K.; Colson, B.; Simon, K.; Mäde, D.; Reiting, R.; Gowik, P.; Grohmann, L. Validation of qualitative PCR methods on the basis of mathematical–statistical modelling of the probability of detection. Accredit. Qual. Assur. 2015, 20, 75–83. [Google Scholar] [CrossRef]
- ISO 5725-2. Accuracy (Trueness and Precision) of Measurement Methods and Results—Part 2: Basic Method for the Determination of Repeatability and Reproducibility of a Standard Measurement Method (ISO 5725-2:1994 Including Technical Corrigendum 1:2002); International Organization for Standardization: Geneva, Switzerland, 2002. [Google Scholar]
- Official Collection of Test Methods, ASU. Statistik—Planung und Statistische Auswertung von Ringversuchen zur Methodenvalidierung; Federal Office of Consumer Protection and Food Safety (BVL), Beuth Verlag: Berlin, Germany, 2006. [Google Scholar]
- PROLab Plus. Software for Planning, Organizing, Performing and Analyzing Interlaboratory Studies. Available online: www.quodata.de (accessed on 25 October 2021).
- ENGL. European Network of GMO Laboratories. Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing. Available online: http://gmo-crl.jrc.ec.europa.eu/guidancedocs.htm (accessed on 31 October 2016).
- Grohmann, L.; Belter, A.; Speck, B.; Goerlich, O.; Guertler, P.; Angers-Loustau, A.; Patak, A. Screening for six genetically modified soybean lines by an event-specific multiplex PCR method: Collaborative trial validation of a novel approach for GMO detection. J. Consum. Prot. Food Saf. 2017, 12, 23–36. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Wang, Q.; He, Y.; Pan, L. Interlaboratory validation of a real-time PCR detection method for bovine- and ovine-derived material. Meat Sci. 2017, 134, 119–123. [Google Scholar] [CrossRef]
- Gondim, C.D.S.; Junqueira, R.G.; de Souza, S.V.C.; Callao, M.P.; Ruisánchez, I. Determining performance parameters in qualitative multivariate methods using probability of detection (POD) curves. Case study: Two common milk adulterants. Talanta 2017, 168, 23–30. [Google Scholar] [CrossRef]
- Wang, Q.; Cai, Y.; He, Y.; Yang, L.; Pan, L. Collaborative ring trial of two real-time PCR assays for the detection of porcine- and chicken-derived material in meat products. PLoS ONE 2018, 13, e0206609. [Google Scholar] [CrossRef]
- Fraiture, M.-A.; Papazova, N.; Roosens, N.H.C. DNA walking strategy to identify unauthorized genetically modified bacteria in microbial fermentation products. Int. J. Food Microbiol. 2021, 337, 108913. [Google Scholar] [CrossRef]
- Zarske, M.; Zagon, J.; Schmolke, S.; Seidler, T.; Braeuning, A. Detection of silkworm (Bombyx mori) and Lepidoptera DNA in feeding stuff by real-time PCR. Food Control 2021, 126, 108059. [Google Scholar] [CrossRef]
- Ali, M.E.; Rahman, M.M.; Hamid, S.B.A.; Mustafa, S.; Bhassu, S.; Hashim, U. Canine-specific PCR assay targeting cytochrome b gene for the detection of dog meat adulteration in commercial frankfurters. Food Anal. Methods 2014, 7, 234–241. [Google Scholar] [CrossRef] [Green Version]
- Cammà, C.; Di Domenico, M.; Monaco, F. Development and validation of fast Real-Time PCR assays for species identification in raw and cooked meat mixtures. Food Control 2012, 23, 400–404. [Google Scholar] [CrossRef]
- Fumière, O.; Dubois, M.; Baeten, V.; Von Holst, C.; Berben, G. Effective PCR detection of animal species in highly processed animal byproducts and compound feeds. Anal. Bioanal. Chem. 2006, 385, 1045–1054. [Google Scholar] [CrossRef]
- Hird, H.; Chisholm, J.; Sanchez, A.; Hernandez, M.; Goodier, R.; Schneede, K.; Boltz, C.; Popping, B. Effect of heat and pressure processing on DNA fragmentation and implications for the detection of meat using a real-time polymerase chain reaction. Food Addit. Contam. 2006, 23, 645–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojas, M.; González, I.; Pavón, M.Á.; Pegels, N.; Hernández, P.E.; García, T.; Martín, R. Application of a real-time PCR assay for the detection of ostrich (Struthio camelus) mislabelling in meat products from the retail market. Food Control 2011, 22, 523–531. [Google Scholar] [CrossRef]
- Druml, B.; Kaltenbrunner, M.; Hochegger, R.; Cichna-Markl, M. A novel reference real-time PCR assay for the relative quantification of (game) meat species in raw and heat-processed food. Food Control 2016, 70, 392–400. [Google Scholar] [CrossRef]
- Uhlig, S.; Lischer, P. Statistically-based performance characteristics in laboratory performance studies. Analyst 1998, 123, 167–172. [Google Scholar] [CrossRef]
- Official Test Method L 00.00-170. Untersuchung von Lebensmitteln. Nachweis von DNA aus Säugetieren und Geflügel in Lebensmitteln Mittels Real-Time PCR auf Basis des Myostatin-Gens; Federal Office of Consumer Protection and Food Safety (BVL), Beuth Verlag: Berlin, Germany, 2020. [Google Scholar]

| Meat Sample | Sample Name | Proportion of Roe Deer (%, w/w) |
|---|---|---|
| 1 | meat mixture 1 | 0 |
| 2 | meat mixture 2 | 1 |
| 3 | meat mixture 3 | 4.9 |
| 4 | meat mixture 4 | 9.5 |
| 5 | meat mixture 5 | 24.8 |
| 6 | meat mixture 6 | 37.2 |
| 7 | meat mixture 7 | 49.4 |
| 8 | meat mixture 8 | 25.1 |
| 9 | meat mixture 9, boiled | 24.9 |
| 10 | model sausage, raw | 21.0 |
| 11 | sausage, brewed | unknown 1 |
| 12 | sausage, raw | unknown 1 |
| Assay | Primer/Probe | Sequence (5′-3′) 1 | Final Concentration [nM] | Reference |
|---|---|---|---|---|
| primer f | TGGCTGCTGCGTGCAGAA | 200 | ||
| roe deer | primer r | TCTAAAATGCTTGGGAACCAGATAT | 200 | [14] |
| probe | FAM-GAAGGGTCTCCGTCTGC-MGBNFQ | 100 | ||
| primer f | TTGTGCARATCCTGAGACTCAT | 200 | ||
| myostatin | primer r | ATACCAGTGCCTGGGTTCAT | 200 | [22] |
| probe | FAM-CCCATGAAAGACGGTACAAGRTATACTG-BHQ1 | 100 |
| Laboratory | Roe Deer Real-Time PCR | Reference Real-Time PCR | ||||
|---|---|---|---|---|---|---|
| Slope | R2 | E (%) | Slope | R2 | E (%) | |
| 1 | −3.5759 | 0.9966 | 90.39 | −3.3792 | 0.9992 | 97.66 |
| 2 | −3.4180 | 0.9963 | 96.14 | −3.3263 | 0.9968 | 99.82 |
| 3 | −3.5573 | 0.9969 | 91.03 | −3.5396 | 0.9985 | 91.65 |
| 4 | −3.6037 | 0.9961 | 89.45 | −3.6082 | 0.9970 | 89.30 |
| 5 | −3.5877 | 0.9942 | 89.99 | −3.3749 | 0.9999 | 97.83 |
| 6 | −3.4349 | 0.9987 | 95.49 | −3.3983 | 0.9937 | 96.91 |
| 7 | −3.4698 | 0.9973 | 94.18 | −3.4947 | 0.9989 | 93.26 |
| 8 | −3.5276 | 0.9970 | 92.08 | −3.3273 | 0.9978 | 99.78 |
| 9 | −3.2937 | 0.9981 | 101.19 | −3.3817 | 0.9996 | 97.56 |
| 10 | −3.5797 | 0.9989 | 90.26 | −3.4860 | 0.9996 | 93.58 |
| 11 | −3.0728 1 | 0.9986 | 111.56 2 | −3.5304 | 0.9615 2 | 91.98 |
| 12 | −3.4141 | 0.9980 | 96.29 | −3.3037 | 0.9994 | 100.77 |
| 13 | −3.4531 | 0.9975 | 94.80 | −3.2621 | 0.9991 | 102.56 |
| 14 | −3.4324 | 0.9986 | 95.59 | −3.3850 | 0.9981 | 97.43 |
| Laboratory | Copy Number/5 µL | ||||||
|---|---|---|---|---|---|---|---|
| 0.1 | 0.5 | 1 | 2 | 5 | 10 | 20 | |
| 1 | 0 | 2 | 5 | 4 | 6 | 6 | 6 |
| 2 | 2 | 4 | 5 | 5 | 6 | 6 | 6 |
| 3 | 0 | 5 | 4 | 6 | 6 | 6 | 6 |
| 4 | 0 | 3 | 4 | 4 | 6 | 6 | 6 |
| 5 | 2 | 4 | 3 | 6 | 6 | 6 | 6 |
| 6 | 0 | 1 | 3 | 4 | 6 | 6 | 6 |
| 7 | 1 | 3 | 6 | 6 | 6 | 6 | 6 |
| 8 | 1 | 2 | 5 | 5 | 6 | 6 | 6 |
| 9 | 0 | 5 | 6 | 5 | 6 | 6 | 6 |
| 10 | 0 | 4 | 5 | 5 | 6 | 6 | 6 |
| 11 | 1 | 1 | 4 | 6 | 6 | 6 | 6 |
| 12 | 1 | 2 | 6 | 5 | 6 | 6 | 6 |
| 13 | 0 | 4 | 3 | 6 | 6 | 6 | 6 |
| 14 | 1 | 2 | 4 | 6 | 6 | 6 | 6 |
| Theoretical Copy Number of the Roe Deer-Specific Target Sequence Per 5 µL | Roe Deer Real-Time PCR | |||
|---|---|---|---|---|
| Number of Positive Tests/Total Number of Tests | Percentage of Positive Tests (%) | pU (%) 1 | pO (%) 2 | |
| 20 | 84/84 | 100.0 | 96.5 | 100.0 |
| 10 | 84/84 | 100.0 | 96.5 | 100.0 |
| 5 | 84/84 | 100.0 | 96.5 | 100.0 |
| 2 | 73/84 | 86.9 | 79.3 | 92.5 |
| 1 | 63/84 | 75.0 | 66.0 | 82.6 |
| 0.5 | 42/84 | 50.0 | 40.5 | 59.5 |
| 0.1 | 9/84 | 10.7 | 5.7 | 18.0 |
| Parameter | Value |
|---|---|
| number of participating laboratories | 14 |
| number of PCR replicates per dilution level | 6 |
| model parameters of the POD curve: | |
| average amplification probability λo | 1.25 |
| 95% confidence interval for the estimated value of λo | 1.05–1.49 |
| estimated value for slope b | 1 |
| laboratory standard deviation σL | 0.15 |
| LOD95% for median laboratory (copy number of the target sequence per 5 µL) | 2.4 |
| Parameter 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | Sample 6 | Sample 7 | Sample 8 | Sample 9 | Sample 10 | Sample 11 | Sample 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| participating labs | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 |
| labs with quantitative results | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 |
| outlier labs | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| labs for determining parameters | 14 | 14 | 14 | 14 | 13 | 14 | 13 | 14 | 14 | 14 | 13 |
| theoretical value (%) | 1.0 | 4.9 | 9.5 | 24.8 | 37.2 | 49.4 | 25.1 | 24.9 | 21.0 | - | - |
| mean ± confidence level (%) | 1.05 ± 0.07 | 5.22 ± 0.42 | 9.90 ± 0.76 | 24.02 ± 3.66 | 37.50 ± 3.19 | 47.40 ± 4.39 | 28.46 ± 1.93 | 132.57 ± 12.91 | 17.72 ± 1.57 | 7.63 ± 0.96 | 8.99 ± 0.86 |
| sR (%) | 0.20 | 0.90 | 1.63 | 7.26 | 6.34 | 9.44 | 3.80 | 26.85 | 3.51 | 1.91 | 1.68 |
| sR rel | 19.50% | 17.26% | 16.46% | 30.22% | 16.91% | 19.91% | 13.35% | 20.25% | 19.83% | 25.08% | 18.71% |
| R (%) | 0.57 | 2.52 | 4.56 | 20.33 | 17.76 | 26.42 | 10.64 | 75.18 | 9.84 | 5.36 | 4.71 |
| R rel | 54.59% | 48.33% | 46.09% | 84.63% | 47.36% | 55.75% | 37.38% | 56.71% | 55.52% | 70.23% | 52.40% |
| sr (%) | 0.19 | 0.56 | 0.98 | 2.93 | 3.29 | 5.71 | 1.88 | 14.36 | 2.35 | 0.83 | 0.82 |
| sr rel | 17.71% | 10.63% | 9.87% | 12.20% | 8.76% | 12.05% | 6.60% | 10.83% | 13.24% | 10.83% | 9.09% |
| r (%) | 0.52 | 1.56 | 2.73 | 8.21 | 9.20 | 16.00 | 5.26 | 40.21 | 6.57 | 2.32 | 2.29 |
| r rel | 49.60% | 29.77% | 27.62% | 34.17% | 24.54% | 33.75% | 18.49% | 30.33% | 37.08% | 30.34% | 25.46% |
| recovery (%) | 105.1 | 106.6 | 104.2 | 96.9 | 100.8 | 95.9 | 113.4 | 532.4 | 84.4 | - 2 | - 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Druml, B.; Uhlig, S.; Simon, K.; Frost, K.; Hettwer, K.; Cichna-Markl, M.; Hochegger, R. Real-Time PCR Assay for the Detection and Quantification of Roe Deer to Detect Food Adulteration—Interlaboratory Validation Involving Laboratories in Austria, Germany, and Switzerland. Foods 2021, 10, 2645. https://doi.org/10.3390/foods10112645
Druml B, Uhlig S, Simon K, Frost K, Hettwer K, Cichna-Markl M, Hochegger R. Real-Time PCR Assay for the Detection and Quantification of Roe Deer to Detect Food Adulteration—Interlaboratory Validation Involving Laboratories in Austria, Germany, and Switzerland. Foods. 2021; 10(11):2645. https://doi.org/10.3390/foods10112645
Chicago/Turabian StyleDruml, Barbara, Steffen Uhlig, Kirsten Simon, Kirstin Frost, Karina Hettwer, Margit Cichna-Markl, and Rupert Hochegger. 2021. "Real-Time PCR Assay for the Detection and Quantification of Roe Deer to Detect Food Adulteration—Interlaboratory Validation Involving Laboratories in Austria, Germany, and Switzerland" Foods 10, no. 11: 2645. https://doi.org/10.3390/foods10112645
APA StyleDruml, B., Uhlig, S., Simon, K., Frost, K., Hettwer, K., Cichna-Markl, M., & Hochegger, R. (2021). Real-Time PCR Assay for the Detection and Quantification of Roe Deer to Detect Food Adulteration—Interlaboratory Validation Involving Laboratories in Austria, Germany, and Switzerland. Foods, 10(11), 2645. https://doi.org/10.3390/foods10112645






