Composition of the Protein Ingredients from Insoluble Oat Byproducts Treated with Food-Grade Enzymes, Such as Amylase, Cellulose/Xylanase, and Protease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Enzymatic Treatments of the Oat Press Cake and Protein Extraction
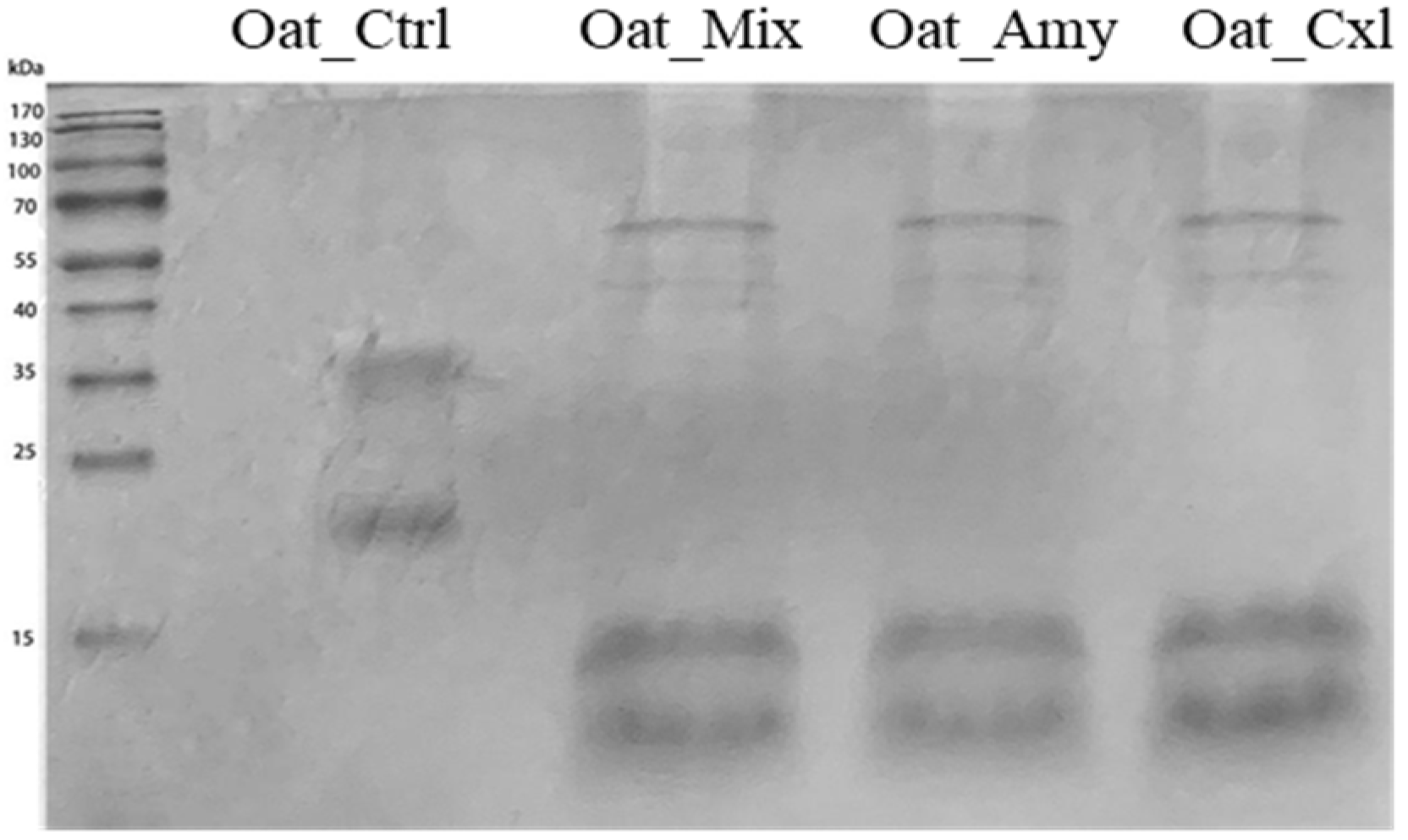
2.3. Analysis by SDS–PAGE and Mass Spectrometry
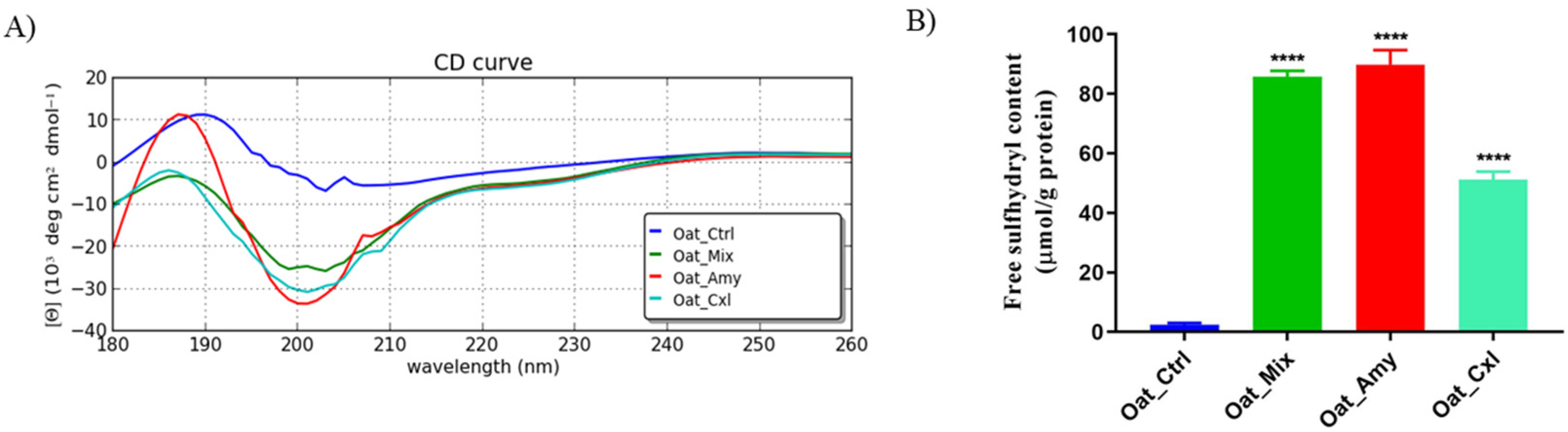
2.4. Circular Dichroism (CD) Spectroscopy
2.5. Degree of Hydrolysis and Free Sulfhydryl Group Determination
2.6. 3 kDa Fractionation, Peptide Content, and LC–MS Analysis
2.7. Extraction of Bound Phenols
2.8. Phenolic Compound Identification and Quantification by MS
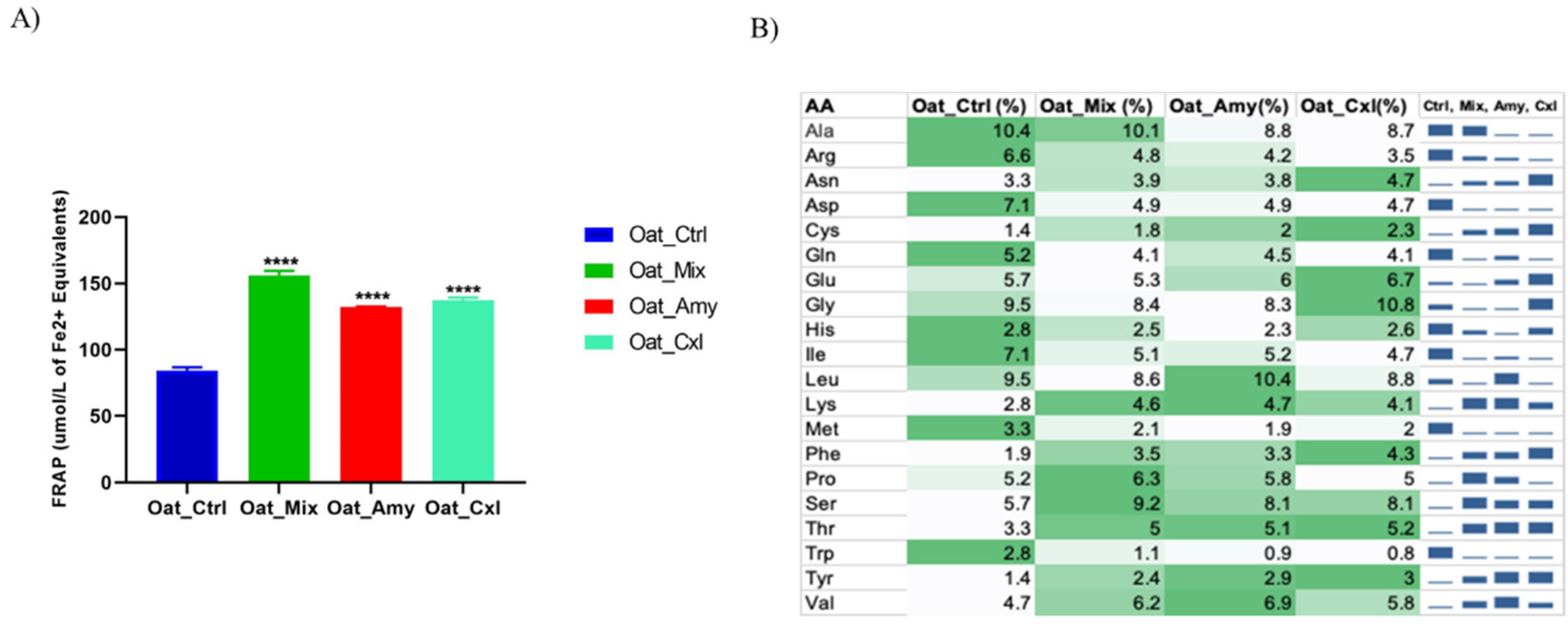
2.9. Radical Scavenging Activity of the 3 kDa Peptide Extracts measured by Ferric Reducing Ability (FRAP) Assay
2.10. Phytic Acid (PA) Quantification
2.11. Total Carbohydrate Quantification
2.12. Statistical Analysis
3. Results and Discussion
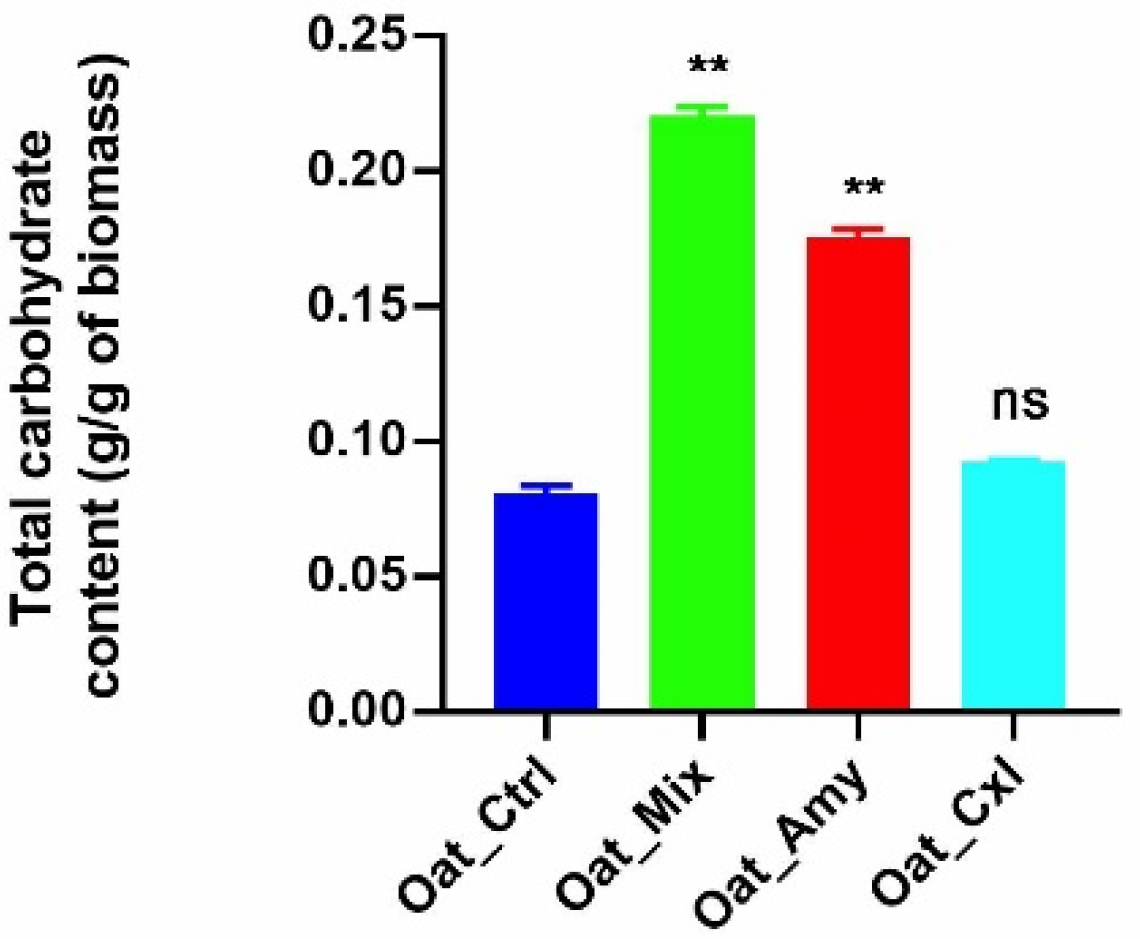
3.1. Effects of the Enzymatic Treatments on Bound Carbohydrates
3.2. Effects of the Enzymatic Treatments on the Primary Structure of the Oat Press Cake Proteins
3.3. Effects of the Enzymatic Treatments on the Secondary and Tertiary Structure of the Proteins
3.4. Analysis of the 3 kDa Peptide Fractions by Liquid Chromatography–Mass Spectrometry (LC–MS)
3.5. Evaluation of the Antioxidant Activity of the Oat Press Cake Proteins and Low Molecular Weight Peptides
3.6. Identification and Quantitation of the Main Phenolic Compounds by HPLC–ESI–MS/MS
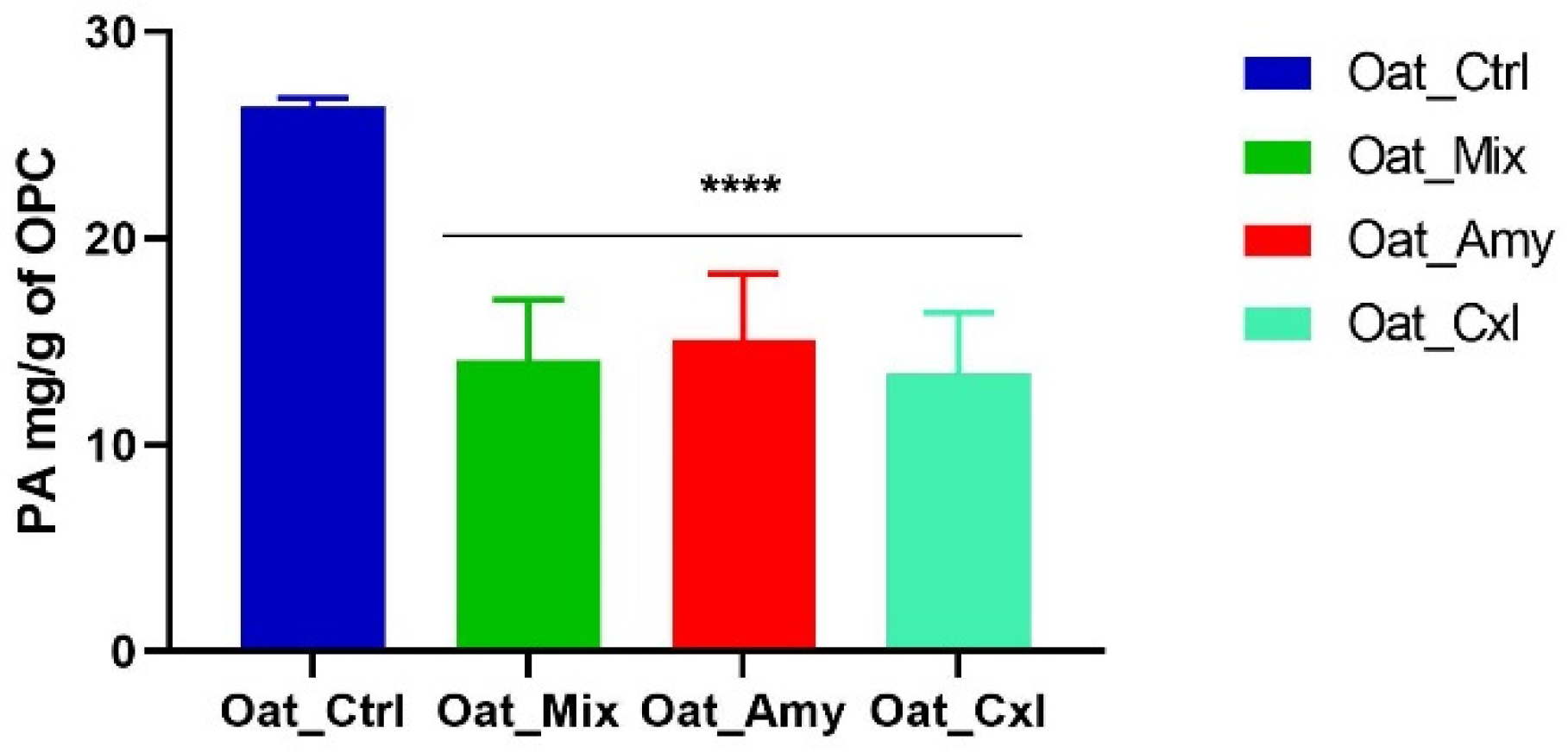
3.7. Effects of Enzymatic Treatments on the Phytic Acid Content
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | amino acid |
| ACN | acetonitrile |
| AMY | amylase |
| AVNs | avenanthramides |
| CD | circular dichroism |
| DTNB | (5,5-dithio-bis-(2-nitrobenzoic acid) |
| FA | formic acid |
| LMW | low molecular weight |
| MIX | amylase/cellulase/protease |
| FeIII-TPTZ | ferric tripyridyltriazine |
| FRAP | ferric reducing ability |
| HPLC–MS/MS | high-performance liquid chromatography–tandem mass spectrometry |
| OPA | o-phthalaldehyde |
| PA | phytic acid |
| SH | free sulfhydryl group |
| Tris–HCl | Tris(hydroxymethyl)aminomethane hydrochloride |
References
- Gangopadhyay, N.; Hossain, M.B.; Rai, D.K.; Brunton, N.P. A Review of Extraction and Analysis of Bioactives in Oat and Barley and Scope for Use of Novel Food Processing Technologies. Molecules 2015, 20, 10884–10909. [Google Scholar] [CrossRef] [PubMed]
- Meydani, M. Potential health benefits of avenanthramides of oats. Nutr. Rev. 2009, 67, 731–735. [Google Scholar] [CrossRef]
- Contreras, M.D.; Lama-Munoz, A.; Gutierrez-Perez, J.M.; Espinola, F.; Moya, M.; Castro, E. Protein extraction from agri-food residues for integration in biorefinery: Potential techniques and current status. Bioresour. Technol. 2019, 280, 459–477. [Google Scholar] [CrossRef]
- Barcelos, M.C.S.; Ramos, C.L.; Kuddus, M.; Rodriguez-Couto, S.; Srivastava, N.; Ramteke, P.W.; Mishra, P.K.; Molina, G. Enzymatic potential for the valorization of agro-industrial by-products. Biotechnol. Lett. 2020, 42, 1799–1827. [Google Scholar] [CrossRef]
- Li, B.B.; Smith, A.B.; Hossain, M.M. Extraction of phenolics from citrus peels II. Enzyme-assisted extraction method. Sep. Purif. Technol. 2006, 48, 189–196. [Google Scholar] [CrossRef]
- Ruviaro, A.R.; Barbosa, P.D.M.; Macedo, G.A. Enzyme-assisted biotransformation increases hesperetin content in citrus juice by-products. Food Res. Int. 2019, 124, 213–221. [Google Scholar] [CrossRef]
- Rommi, K.; Holopainen, U.; Pohjola, S.; Hakala, T.K.; Lantto, R.; Poutanen, K.; Nordlund, E. Impact of Particle Size Reduction and Carbohydrate-Hydrolyzing Enzyme Treatment on Protein Recovery from Rapeseed (Brassica rapa L.) Press Cake. Food Bioprocess Technol. 2015, 8, 2392–2399. [Google Scholar] [CrossRef]
- Jodayree, S.; Smith, J.C.; Tsopmo, A. Use of carbohydrase to enhance protein extraction efficiency and antioxidative properties of oat bran protein hydrolysates. Food Res. Int. 2012, 46, 69–75. [Google Scholar] [CrossRef]
- Rosset, M.; Acquaro, V.R.; Beleia, A.D. Protein Extraction from Defatted Soybean Flour with Viscozyme L Pretreatment. J. Food Process. Preserv. 2014, 38, 784–790. [Google Scholar] [CrossRef]
- Alrahmany, R.; Tsopmo, A. Role of carbohydrases on the release of reducing sugar, total phenolics and on antioxidant properties of oat bran. Food Chem. 2012, 132, 413–418. [Google Scholar] [CrossRef]
- Gligor, O.; Mocan, A.; Moldovan, C.; Locatelli, M.; Crisan, G.; Ferreira, I. Enzyme-assisted extractions of polyphenols—A comprehensive review. Trends Food Sci. Technol. 2019, 88, 302–315. [Google Scholar] [CrossRef]
- Puri, M.; Sharma, D.; Barrow, C.J. Enzyme-assisted extraction of bioactives from plants. Trends Biotechnol. 2012, 30, 37–44. [Google Scholar] [CrossRef]
- Aiello, G.; Lammi, C.; Boschin, G.; Zanoni, C.; Arnoldi, A. Exploration of Potentially Bioactive Peptides Generated from the Enzymatic Hydrolysis of Hempseed Proteins. J. Agric. Food Chem. 2017, 65, 10174–10184. [Google Scholar] [CrossRef]
- Aiello, G.; Fasoli, E.; Boschin, G.; Lammi, C.; Zanoni, C.; Citterio, A.; Arnoldi, A. Proteomic characterization of hempseed (Cannabis sativa L.). J. Proteom. 2016, 147, 187–196. [Google Scholar] [CrossRef]
- Verardo, V.; Serea, C.; Segal, R.; Caboni, M.F. Free and bound minor polar compounds in oats: Different extraction methods and analytical determinations. J. Cereal Sci. 2011, 54, 211–217. [Google Scholar] [CrossRef]
- Raussens, V.; Ruysschaert, J.M.; Goormaghtigh, E. Protein concentration is not an absolute prerequisite for the determination of secondary structure from circular dichroism spectra: A new scaling method. Anal. Biochem. 2003, 319, 114–121. [Google Scholar] [CrossRef]
- Nielsen, P.M.; Petersen, D.; Dambmann, C. Improved method for determining food protein degree of hydrolysis. J. Food Sci. 2001, 66, 642–646. [Google Scholar] [CrossRef]
- Zhao, G.L.; Liu, Y.; Zhao, M.M.; Ren, J.Y.; Yang, B. Enzymatic hydrolysis and their effects on conformational and functional properties of peanut protein isolate. Food Chem. 2011, 127, 1438–1443. [Google Scholar] [CrossRef]
- Rohm, H.; Tschager, E.; Jaros, D. Determination of proteolysis in Swiss cheese: Comparison of the Kjeldahl method and a spectrophotometric OPA assay. Food Sci. Technol.-Lebensm.-Wiss.-Technol. 1996, 29, 191–194. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Shang, C.; Maroof, M.A.S.; Biyashev, R.M.; Grabau, E.A.; Kwanyuen, P.; Burton, J.W.; Buss, G.R. A modified colorimetric method for phytic acid analysis in soybean. Crop Sci. 2007, 47, 1797–1803. [Google Scholar] [CrossRef] [Green Version]
- Mecozzi, M. Estimation of total carbohydrate amount in environmental samples by the phenol-sulphuric acid method assisted by multivariate calibration. Chemom. Intell. Lab. Syst. 2005, 79, 84–90. [Google Scholar] [CrossRef]
- Boukid, F. Oat proteins as emerging ingredients for food formulation: Where we stand? Eur. Food Res. Technol. 2021, 247, 535–544. [Google Scholar] [CrossRef]
- Gazza, L.; Taddei, F.; Conti, S.; Gazzelloni, G.; Muccilli, V.; Janni, M.; D’Ovidio, R.; Alfieri, M.; Redaelli, R.; Pogna, N.E. Biochemical and molecular characterization of Avena indolines and their role in kernel texture. Mol. Genet. Genom. 2015, 290, 39–54. [Google Scholar] [CrossRef]
- Kaukonen, O.; Sontag-Strohm, T.; Salovaara, H.; Lampi, A.M.; Sibakov, J.; Loponen, J. Foaming of Differently Processed Oats: Role of Nonpolar Lipids and Tryptophanin Proteins. Cereal Chem. 2011, 88, 239–244. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Bulyaki, E.; Kun, J.; Moussong, E.; Lee, Y.H.; Goto, Y.; Refregiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef]
- Wen, C.T.; Zhang, J.X.; Zhou, J.; Duan, Y.Q.; Zhang, H.H.; Ma, H.L. Effects of slit divergent ultrasound and enzymatic treatment on the structure and antioxidant activity of arrowhead protein. Ultrason. Sonochem. 2018, 49, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Stadnik, J.; Dolatowski, Z.J. Changes in selected parameters related to proteolysis during ageing of dry-cured pork loins inoculated with probiotics. Food Chem. 2013, 139, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.B.; Zhu, W.D.; Yi, J.H.; Liu, N.; Cao, Y.G.; Lu, J.L.; Decker, E.A.; McClements, D.J. Effects of sonication on the physicochemical and functional properties of walnut protein isolate. Food Res. Int. 2018, 106, 853–861. [Google Scholar] [CrossRef]
- Shahidi, F.; Ambigaipalan, P. Phenolics and polyphenolics in foods, beverages and spices: Antioxidant activity and health effects—A review. J. Funct. Foods 2015, 18, 820–897. [Google Scholar] [CrossRef]
- Lammi, C.; Aiello, G.; Boschin, G.; Arnoldi, A. Multifunctional peptides for the prevention of cardiovascular disease: A new concept in the area of bioactive food-derived peptides. J. Funct. Foods 2019, 55, 135–145. [Google Scholar] [CrossRef]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.C.; Esfandi, R.; Willmore, W.G.; Tsopmo, A. Antioxidant Activity of Oat Proteins Derived Peptides in Stressed Hepatic HepG2 Cells. Antioxidants 2016, 5, 39. [Google Scholar] [CrossRef] [Green Version]
- Nwachukwu, I.D.; Aluko, R.E. Antioxidant Properties of Flaxseed Protein Hydrolysates: Influence of Hydrolytic Enzyme Concentration and Peptide Size. J. Am. Oil Chem. Soc. 2018, 95, 1105–1118. [Google Scholar] [CrossRef]
- Nwachukwu, I.D.; Aluko, R.E. Structural and functional properties of food protein-derived antioxidant peptides. J. Food Biochem. 2019, 43, e12761. [Google Scholar] [CrossRef] [Green Version]
- Muley, A.B.; Pandit, A.B.; Singhal, R.S.; Dalvi, S.G. Production of biologically active peptides by hydrolysis of whey protein isolates using hydrodynamic cavitation. Ultrason. Sonochem. 2021, 71, 105385. [Google Scholar] [CrossRef]
- Chang, O.K.; Ha, G.E.; Han, G.S.; Seol, K.H.; Kim, H.W.; Jeong, S.G.; Oh, M.H.; Park, B.Y.; Ham, J.S. Novel Antioxidant Peptide Derived from the Ultrafiltrate of Ovomucin Hydrolysate. J. Agric. Food Chem. 2013, 61, 7294–7300. [Google Scholar] [CrossRef]
- Debelo, H.; Li, M.; Ferruzzi, M.G. Processing influences on food polyphenol profiles and biological activity. Curr. Opin. Food Sci. 2020, 32, 90–102. [Google Scholar] [CrossRef]
- Wang, T.; He, F.L.; Chen, G.B. Improving bioaccessibility and bioavailability of phenolic compounds in cereal grains through processing technologies: A concise review. J. Funct. Foods 2014, 7, 101–111. [Google Scholar] [CrossRef]
- Quan, T.H.; Benjakul, S.; Sae-leaw, T.; Balange, A.K.; Maqsood, S. Protein-polyphenol conjugates: Antioxidant property, functionalities and their applications. Trends Food Sci. Technol. 2019, 91, 507–517. [Google Scholar] [CrossRef]
- Mattila, P.; Pihlava, J.M.; Hellstrom, J. Contents of phenolic acids, alkyl- and alkenylresorcinols, and avenanthramides in commercial grain products. J. Agric. Food Chem. 2005, 53, 8290–8295. [Google Scholar] [CrossRef]
- Xu, J.G.; Tian, C.R.; Hu, Q.P.; Luo, J.Y.; Wang, X.D.; Tian, X.D. Dynamic Changes in Phenolic Compounds and Antioxidant Activity in Oats (Avena nuda L.) during Steeping and Germination. J. Agric. Food Chem. 2009, 57, 10392–10398. [Google Scholar] [CrossRef] [PubMed]
- Multari, S.; Pihlava, J.M.; Ollennu-Chuasam, P.; Hietaniemi, V.; Yang, B.R.; Suomela, J.P. Identification and Quantification of Avenanthramides and Free and Bound Phenolic Acids in Eight Cultivars of Husked Oat (Avena sativa L) from Finland. J. Agric. Food Chem. 2018, 66, 2900–2908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soycan, G.; Schar, M.Y.; Kristek, A.; Boberska, J.; Alsharif, S.N.S.; Corona, G.; Shewry, P.R.; Spencer, J.P.E. Composition and content of phenolic acids and avenanthramides in commercial oat products: Are oats an important polyphenol source for consumers? Food Chem.-X 2019, 3, 100047. [Google Scholar] [CrossRef]
- Chen, D.F.; Shi, J.L.; Hu, X.Z. Enhancement of polyphenol content and antioxidant capacity of oat (Avena nuda L.) bran by cellulase treatment. Appl. Biol. Chem. 2016, 59, 397–403. [Google Scholar] [CrossRef]
- Jagr, M.; Dvoracek, V.; Cepkova, P.H.; Dolezalova, J. Comprehensive analysis of oat avenanthramides using hybrid quadrupole-Orbitrap mass spectrometry: Possible detection of new compounds. Rapid Commun. Mass Spectrom. 2020, 34, e8718. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.L.; Zheng, J.B.; Wang, X.D.; Chen, F.S. Effects of household cooking processes on mineral, vitamin B, and phytic acid contents and mineral bioaccessibility in rice. Food Chem. 2019, 280, 59–64. [Google Scholar] [CrossRef]
- Sivakumar, V.; Swaminathan, G.; Rao, P.G. Use of ultrasound in soaking for improved efficiency. J. Soc. Leather Technol. Chem. 2004, 88, 249–251. [Google Scholar]
| Component | Percentage |
|---|---|
| Moisture | 64.17 ± 0.08 |
| Protein | 32.42 ± 0.45 * |
| Lipids | 7.79 ± 0.03 * |
| Insoluble dietary fibre | 22.97 ± 0.15 * |
| Soluble dietary fibre | 3.19 ± 0.02 * |
| Starch | 27.32 ± 0.20 * |
| Enzyme | Activity, U/g | Organism of Origin | Optimal pH | Optimal Temperature, °C |
|---|---|---|---|---|
| α-Amylase | >1400 | Bacillus licheniformis | 5.5–7.5 | 70–85 |
| Cellulase/xylanase mixture | >45,000 cellulase >34,000 xylanase | Trichoderma reesei | 5.0–6.5 | 40–60 |
| Protease | 116,350 | Aspergillus oryzae | 6.0–7.5 | 30–50 |
| Distinct Summed MS/MS Search Score | % AA Coverage | Total Spectrum Intensity | Protein MW | Protein pI | Accession (#)(a) | Protein Name |
|---|---|---|---|---|---|---|
| Oat_Ctrl | ||||||
| 27.86 | 4.8 | 6.61 × 108 | 58,999.9 | 9.22 | P12615 | 12S seed storage globulin 1 |
| 27.86 | 7.9 | 6.61 × 108 | 35,779.4 | 10.2 | Q38781 | Oat storage protein 12S globulin (Fragment) |
| 18.96 | 2.3 | 6.55 × 108 | 58,566.4 | 8.8 | O49258 | 12S globulin |
| 18.96 | 5.6 | 6.55 × 108 | 24,685.5 | 7.94 | P27919 | Avenin |
| 28.39 | 4.7 | 1.27 × 108 | 59,804 | 9.52 | Q38780 | 11S globulin |
| 5.73 | 4.1 | 1.19 × 108 | 59,773.6 | 5.38 | A0A4Y5UJ50 | 4-coumarate:CoA ligase |
| 6.26 | 9.4 | 6.95 × 108 | 23,329.1 | 11.17 | A0A3G1AXD2 | Ribosomal protein S4 |
| 7.26 | 4.2 | 6.11 × 107 | 53,845.5 | 5.16 | A0A3G1AWG0 | ATP synthase subunit beta |
| 5.58 | 8.2 | 2.79 × 107 | 28,805.3 | 5.98 | I4IY74 | Pollen allergen Ave s 5 (Isoallergen A) |
| 6.65 | 1.4 | 2.17 × 107 | 1,221,607 | 9.18 | A0A3G1AUJ8 | DNA-directed RNA polymerase subunit beta |
| 5.77 | 17.7 | 1.55 × 107 | 10,773.6 | 11.07 | A0A3G1AU31 | 30S ribosomal protein S15 |
| 6.05 | 4.3 | 1.30 × 107 | 50,203.8 | 8.69 | Q941N4 | Receptor kinase |
| Oat_Mix, Amy, Cxl | ||||||
| 5.55 | 14.6 | 1.15 × 109 | 16,544 | 8.3 | A0A1B2LQF1 | Avena alpha amylase trypsin inhibitor |
| 10.1 | 5.5 | 1.05 × 109 | 78,923 | 9.03 | A0A3G1ATL7 | DNA-directed RNA polymerase subunit gamma |
| 9.8 | 12.2 | 9.76 × 108 | 30,425 | 5.28 | I4IY75 | Pollen allergen Ave s 5 (Isoallergen B) |
| 15.5 | 4.2 | 6.07 × 108 | 100,456 | 7.61 | A0A482JYP4 | Phototropin-like protein |
| 11.78 | 6.1 | 5.38 × 108 | 59,614 | 5.39 | P54411 | T-complex protein 1 subunit epsilon |
| 4.57 | 3.9 | 4.67 × 108 | 58,836 | 8.14 | F5B4I6 | Non-specific serine/threonine protein kinase |
| 6.22 | 7.6 | 2.74 × 108 | 32,980 | 8.86 | Q7XXP0 | Hydroxyanthranilate hydroxycinnamoyltransferase 4 (Fragm) |
| 5.31 | 1.8 | 1.75 × 108 | 96,836 | 5.66 | G1JSL5 | Lipoxygenase |
| 15.08 | 16.6 | 1.66 × 108 | 41260 | 6.6 | D5L0B2 | Putative 2-oxoglutarate dependent dioxygenase |
| 21.65 | 5.2 | 1.59 × 108 | 170,146 | 7.26 | A0A3G1AXC3 | RNA polymerase beta subunit |
| 12.49 | 8.6 | 1.31 × 108 | 53,847 | 5.16 | A0A3G1AWG0 | ATP synthase subunit beta |
| 15.26 | 4.2 | 1.26 × 108 | 126,405 | 5.75 | P06594 | Phytochrome A type 4 |
| 11.29 | 9.6 | 1.14 × 108 | 49,110 | 5.66 | A0A481SVJ0 | Phenylalanine ammonia lyase II (Fragm) |
| 5.82 | 11.9 | 1.10 × 108 | 16,508 | 8.71 | R4I3I8 | Vromindoline 3 |
| 5.82 | 11.9 | 1.10 × 108 | 16,482 | 8.34 | A7U440 | Tryptophanin |
| 13.9 | 12.5 | 9.08 × 107 | 53,822 | 6.17 | Q43380 | Non-specific serine/threonine protein kinase |
| 7.36 | 5.9 | 8.75 × 107 | 67,169 | 6.06 | P22220 | Arginine decarboxylase |
| 5.33 | 6.3 | 8.38 × 107 | 35,780 | 10.13 | Q38781 | Oat storage protein 12S globulin (Fragm) |
| 13.55 | 14.9 | 7.25 × 107 | 22,640 | 5.5 | A0A2P0ZEN0 | Ribulose bisphosphate carboxylase large chain (Fragm) |
| 13.08 | 5.9 | 3.89 × 107 | 112,385 | 6.48 | Q38766 | Glycine cleavage system P protein |
| 6.91 | 10.8 | 3.73 × 107 | 20,706 | 6.13 | A0A0R6HRG0 | Ribulose bisphosphate carboxylase large chain (Fragm) |
| 5.29 | 3.8 | 2.01 × 107 | 42,153 | 9.01 | Q9LLD7 | Fructose-bisphosphate aldolase |
| 6.83 | 3.4 | 1.82 × 107 | 65,678 | 6.03 | Q38786 | Avenacosidase 1 |
| 3.71 | 1.2 | 1.69 × 107 | 105,789 | 8.63 | A0A3Q8R3E1 | Cellulose synthase-like CslF6 |
| 3.68 | 13.9 | 1.39 × 107 | 18,766 | 7.84 | Q071L4 | Aluminum-activated malate transporter (Fragm) |
| 3.46 | 20.9 | 1.05 × 107 | 9356 | 8.8 | A0A2L0U0E7 | Defensin 16 |
| 7.28 | 4.6 | 4.29 × 106 | 53,776 | 9.36 | O49257 | 12S globulin |
| 7.28 | 4.1 | 4.69 × 106 | 59,811 | 9.14 | Q38780 | 11S globulin |
| α-Helix (%) | β-Sheet (%) | Turn (%) | Others (%) | |
|---|---|---|---|---|
| Oat_Ctrl | 22.2 | 8.5 | 32.7 | 36.6 |
| Oat_Mix | 8.9 | 20.3 | 13.7 | 57.1 |
| Oat_Amy | 3.7 | 28.0 | 7.2 | 61.2 |
| Oat_Cxl | 5.9 | 28.1 | 7.4 | 58.6 |
| Spectrum Intensity | Peptide Sequence | m/z (Da) | % AA Coverage | MH+ (Da) | Peptide pI | Protein MW (Da) | Accession # | Protein Name |
|---|---|---|---|---|---|---|---|---|
| Mix | ||||||||
| 1.40 × 108 | SQQGPVEHQAYQPIQS | 599.57 | 1.2 | 1796.9 | 5.22 | 58,674.5 | P14812 | 12S seed storage globulin 2 |
| 4.13 × 107 | ALGISQQAAQRIQSQNDQRGEI | 804.27 | 1.3 | 2411.2 | 6.12 | 59,404.6 | Q38780 | 11S globulin |
| 4.09 × 107 | DLGADVR | 746.06 | 3.0 | 745.4 | 4.21 | 65,692.1 | Q9ZP27 | Avenacosidase 2 |
| 3.48 × 107 | YQPIQSQEGQSTQYQVGQSTQ | 795.80 | 3.1 | 2385.1 | 4 | 58,224.1 | O49258 | 12Sglobulin |
| 1.44 × 107 | QQSEIMKQVHVAQTLPSK | 684.75 | 2.5 | 2052.1 | 8.6 | 15,927.3 | R4I3I8 | Vromindoline 3 |
| 5.96 × 106 | TNPNSMVSHIAGKSSILRALPVDVLAN | 935.66 | 1.5 | 2804.5 | 8.44 | 58,224.1 | O49258 | 12S globulin |
| 5.93 × 106 | KGTLDGGINHEGIQYYNDL | 703.26 | 1.8 | 2107.0 | 4.54 | 65,039.4 | Q38786 | Avenacosidase 1 |
| 5.50 × 106 | FLVQQCSPVAAVSFLRSQILQQSSCQ | 956.79 | 1.9 | 2867.5 | 8.07 | 24,076.7 | L0L845 | Gliadin-like avenin |
| 2.58 × 106 | NNRGEEFGAFTPKFAQTGSQSYRTRE | 993.74 | 2.3 | 2978.4 | 8.59 | 35,722.3 | Q38781 | Oat storage protein 12S globulin (Fragm) |
| Amy | ||||||||
| 1.82 × 107 | LQQVTQGIFQPQMQGQIEGMRAFA | 903.08 | 3.2 | 2706.4 | 6 | 25,275.1 | P80356 | Avenin-3 |
| 1.61 × 107 | MAQLFGQSSTPWQSSRQGG | 685.03 | 1.0 | 2053.0 | 9.5 | 61,861.4 | Q38779 | 11S globulin |
| 1.17 × 107 | QQQQQQQPFVQQQQMF | 683.35 | 1.1 | 2049.0 | 5.52 | 24,012.3 | L0L6K1 | Gliadin-like avenin |
| 9.98 × 106 | LQLQQQVFQPQLQQQVFQPQL | 855.81 | 2.8 | 2566.4 | 5.52 | 25,471.1 | Q09072 | Avenin |
| 8.33 × 106 | TFNEPHSFCGLGYGTGLHAPGAR | 796.91 | 2.1 | 2389.1 | 6.61 | 65,039.4 | Q38786 | Avenacosidase 1 |
| 7.93 × 105 | YFDEQNEQFRCTG | 546.07 | 3.5 | 1636.7 | 4.14 | 61,861.4 | Q38779 | 11S globulin |
| Cxl | ||||||||
| 1.38 × 107 | LQALPAMCDVYVPPHCPVATTPXGF | 918.14 | 2.0 | 2753.4 | 5.08 | 24,076.7 | L0L845 | Gliadin-like avenin |
| Ctrl 6.73 × 106 | KIQSQNDQRGEIIRV | 595.10 | 1.0 | 1783.9 | 8.75 | 58,674.5 | O49258 | 12S globulin |
| Polyphenols (m/z) | Oat_Ctrl (Mean ± SD) (µg/g DW) | Oat_Mix (Mean ± SD) (µg/g DW) | Oat_Amy (Mean ± SD) (µg/g DW) | Oat_Cxl (Mean ± SD) (µg/g DW) |
| Cinnamic acid (149.2) | 12.1 ± 0.4 | 4.7 ± 0.7 (a) | 5.1 ± 0.4 (a) | 6.6 ± 0.2 (a) |
| p-Coumaric acid (165.2) | 63.1 ± 4.1 | 20.1 ± 8.9 (c) | 60.4 ± 0.1 ns | 16.4 ± 1.2 (a) |
| Vanillic acid (169.1) | 407.6 ± 60.7 | 94.3 ± 23.8 c) | 228.5 ± 78.1 (d) | 178.0 ± 21.7 (c) |
| Gallic acid (171.1) | 2894.3 ± 435.5 | 806.4 ± 4.5 (c) | 1241.1 ± 114.6 (c) | 579.7 ± 60.9 (b) |
| Caffeic acid (181.2) | 4.7 ± 0.8 | 262.2 ± 30.9 (b) | 18.5 ± 2.0 (b) | 265.3 ± 37.0 (b) |
| Ferulic acid (195.2) | 1.9 ± 0.1 | 0.7 ± 0.1 (b) | 0.7 ± 0.1 (b) | 0.99 ± 0.03 (b) |
| AVN (m/z) | Oat_Ctrl (Mean ± SD) (ppb) | Oat_Mix (b) (Mean ± SD) (ppb) | Oat_Amy (a) (Mean ± SD) (ppb) | Oat_Cxl (c) (Mean ± SD) (ppb) |
| AVN A (300.1) | 6.4 ± 0.8 | 14.8 ± 2.8 (c) | 9.6 ± 0.9 (d) | 11.1 ± 4 ns |
| AVN B (330.1) | 45.6 ± 8.8 | 12.8 ± 1.2 (d) | 4.3 ± 0.3 (d) | 6.0 ± 0.8 (d) |
| AVN C (316.1) | 7.9 ± 2.2 | 10.9 ± 3.5 ns | 5.6 ± 0.8 ns | 26.4 ± 0.5 (c) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aiello, G.; Li, Y.; Xu, R.; Boschin, G.; Juodeikiene, G.; Arnoldi, A. Composition of the Protein Ingredients from Insoluble Oat Byproducts Treated with Food-Grade Enzymes, Such as Amylase, Cellulose/Xylanase, and Protease. Foods 2021, 10, 2695. https://doi.org/10.3390/foods10112695
Aiello G, Li Y, Xu R, Boschin G, Juodeikiene G, Arnoldi A. Composition of the Protein Ingredients from Insoluble Oat Byproducts Treated with Food-Grade Enzymes, Such as Amylase, Cellulose/Xylanase, and Protease. Foods. 2021; 10(11):2695. https://doi.org/10.3390/foods10112695
Chicago/Turabian StyleAiello, Gilda, Yuchen Li, Ruoxian Xu, Giovanna Boschin, Grazina Juodeikiene, and Anna Arnoldi. 2021. "Composition of the Protein Ingredients from Insoluble Oat Byproducts Treated with Food-Grade Enzymes, Such as Amylase, Cellulose/Xylanase, and Protease" Foods 10, no. 11: 2695. https://doi.org/10.3390/foods10112695
APA StyleAiello, G., Li, Y., Xu, R., Boschin, G., Juodeikiene, G., & Arnoldi, A. (2021). Composition of the Protein Ingredients from Insoluble Oat Byproducts Treated with Food-Grade Enzymes, Such as Amylase, Cellulose/Xylanase, and Protease. Foods, 10(11), 2695. https://doi.org/10.3390/foods10112695









