A Novel Neutral and Mesophilic β-Glucosidase from Coral Microorganisms for Efficient Preparation of Gentiooligosaccharides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Vectors, and Reagents
2.2. Coral Sample Collection, Activity Screening, DNA Extraction, and High-Throughput Sequencing
2.3. The Recombinant β-Glucosidase Blg163 Expression and Purification
2.4. Activity Assay of β-Glucosidase
2.5. Characterization of Purified Blg163
2.6. Gentiooligosaccharide Production via β-Glucosidase-Mediated Reverse Hydrolysis with Glucose
2.7. Gentiooligosaccharide Synthesis Using Glucose and Cellobiose via Transglycosylation
2.8. Thin-Layer Chromatography (TLC) and High-Performance Liquid Chromatography Analysis of Gentiooligosaccharides Produced by Blg163
2.9. Bioinformatic Analysis and Nucleotide Sequence Submission
3. Results
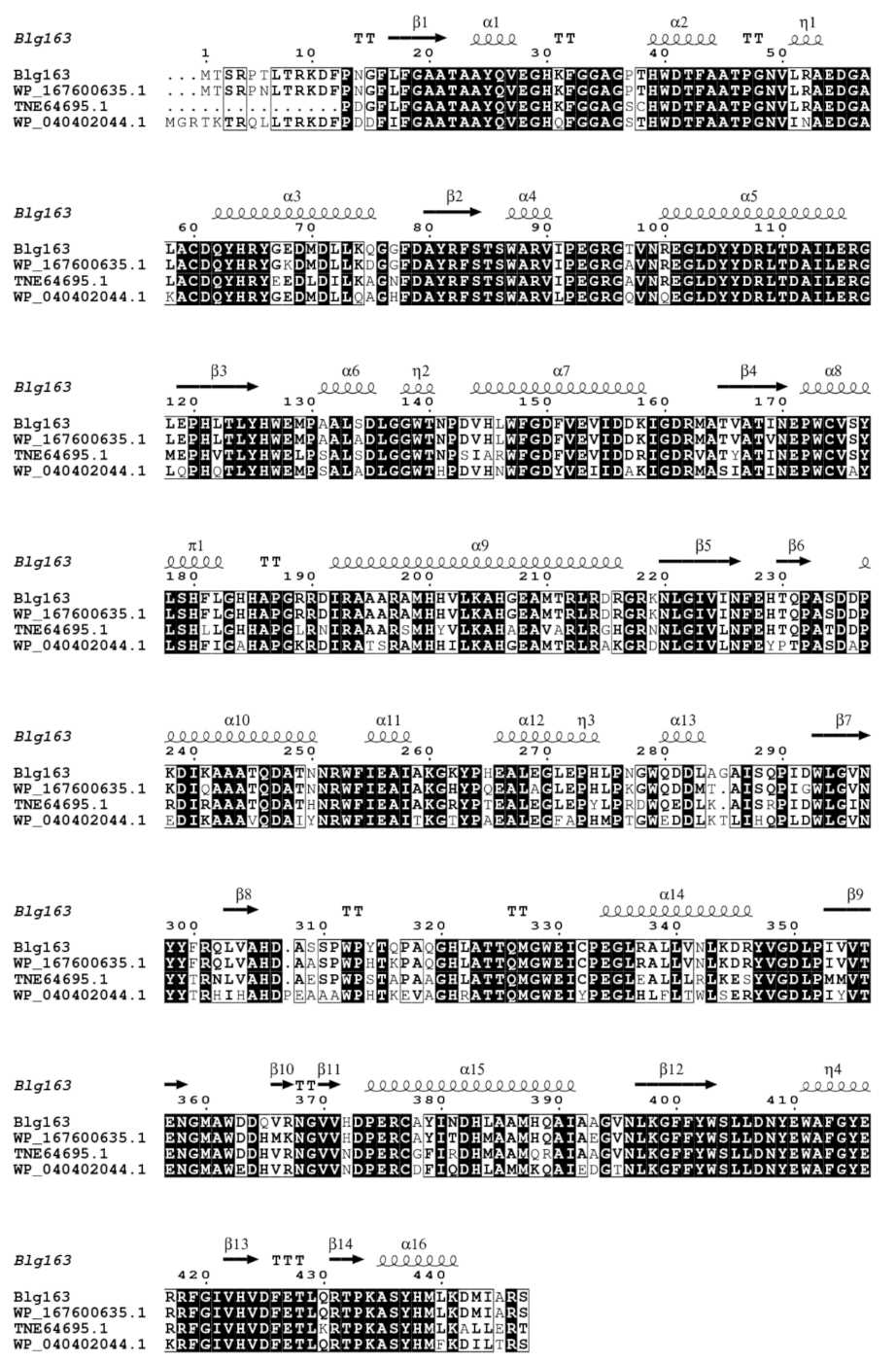
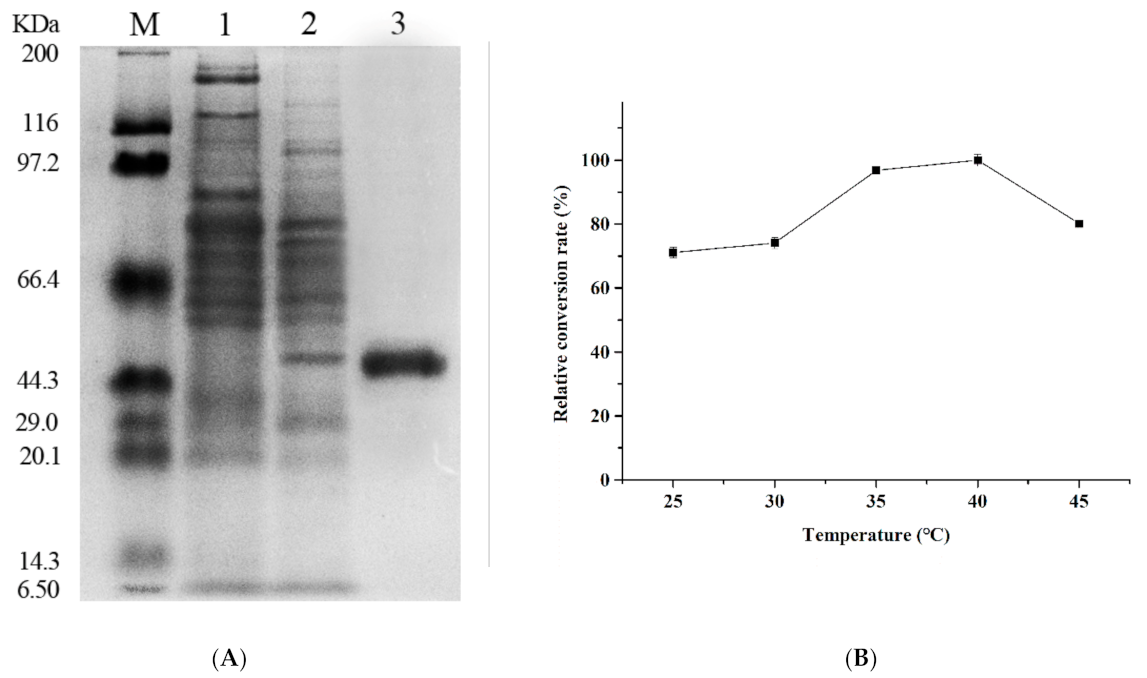
3.1. Cloning, Expression, and Purification of β-Glucosidase
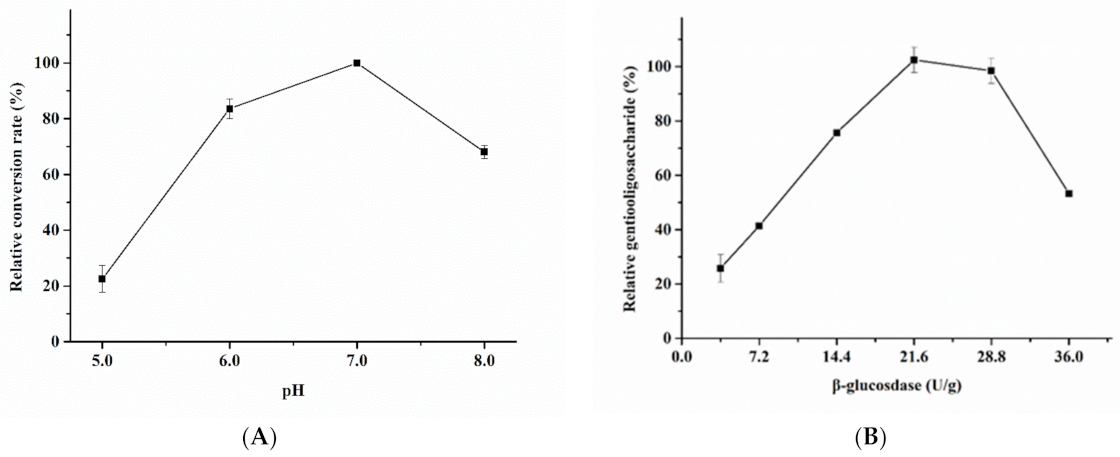
3.2. Enzymatic Properties of Recombinant β-Glucosidase
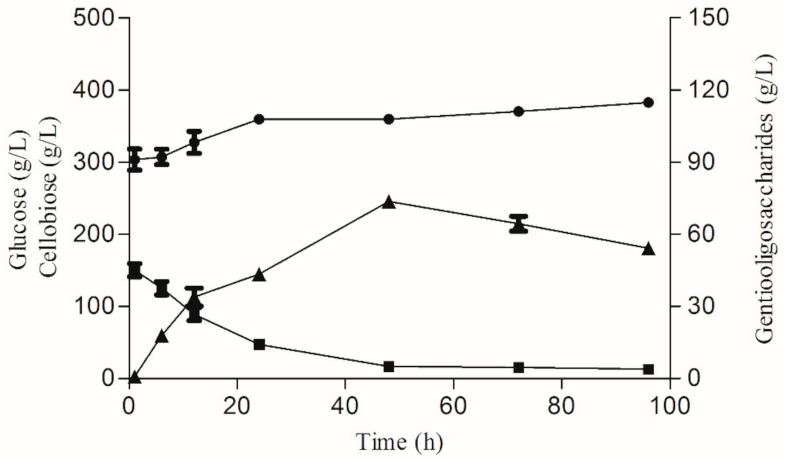
3.3. Gentiooligosaccharide Production from Cellobiose and Glucose
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kim, T.Y.; Lee, D.S.; Shin, H.J. Gentiobiose synthesis from glucose using recombinant β-glucosidase from Thermus caldophilus GK24. Biotechnol. Bioprocess Eng. 2003, 8, 210–212. [Google Scholar] [CrossRef]
- Smaali, M.I.; Michaud, N.; Marzouki, N.; Legoy, M.D.; Maugard, T. Comparison of two β-glucosidases for the enzymatic synthesis of β-(1-6)-β-(1-3)-gluco-oligosaccharides. Biotechnol. Lett. 2004, 26, 675–679. [Google Scholar] [CrossRef] [PubMed]
- Rycroft, C.E.; Jones, M.R.; Gibson, G.R.; Rastall, R.A. Fermentation properties of gentiooligosaccharides. Lett. Appl. Microbiol. 2001, 32, 156–161. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Luo, J.; Li, X.; Pinelo, M. Enzyme membrane reactors for production of oligosaccharides: A review on the interdependence between enzyme reaction and membrane separation. Sep. Pur. Technol. 2020, 243, 116840. [Google Scholar] [CrossRef]
- Gibson, G.R.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wu, J.; Chen, S. Preparation of gentiooligosaccharides using Trichoderma viride beta-glucosidase. Food Chem. 2018, 248, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Hattori, T.; Uno, S.; Murata, T.; Usui, T. Enzymatic synthesis of gentiooligosaccharides by transglycosylation with beta-glycosidases from Penicillium multicolor. Carbohydr. Res. 2009, 344, 972–978. [Google Scholar] [CrossRef] [Green Version]
- Kono, H.; Kawano, S.; Tajima, K.; Erata, T.; Takai, M. Structural analyses of new tri- and tetrasaccharides produced from di-saccharides by transglycosylation of purified Trichoderma viride β-glucosidase. Glycoconj. J. 1999, 16, 415–423. [Google Scholar] [CrossRef]
- Park, T.-H.; Choi, K.-W.; Park, C.-S.; Lee, S.-B.; Kang, H.-Y.; Shon, K.-J.; Park, J.-S.; Cha, J. Substrate specificity and transglycosylation catalyzed by a thermostable β-glucosidase from marine hyperthermophile Thermotoga neapolitana. Appl. Microbiol. Biotechnol. 2005, 69, 411–422. [Google Scholar] [CrossRef]
- Chang, J.; Park, I.H.; Lee, Y.S.; Ahn, S.C.; Zhou, Y.; Choi, Y.L. Cloning, expression, and characterization of β-glucosidase from Exiguobacterium sp. DAU5 and transglycosylation activity. Biotechnol. Bioprocess Eng. 2011, 16, 97–106. [Google Scholar] [CrossRef]
- Qin, Y.; Zhang, Y.; He, H.; Zhu, J.; Chen, G.; Li, W.; Liang, Z. Screening and identification of a fungal beta-glucosidase and the enzymatic synthesis of gentiooligosaccharide. Appl. Biochem. Biotechnol. 2011, 163, 1012–1029. [Google Scholar] [CrossRef]
- Yang, S.; Hua, C.; Yan, Q.; Li, Y.; Jiang, Z. Biochemical properties of a novel glycoside hydrolase family 1 beta-glucosidase (PtBglu1) from Paecilomyces thermophila expressed in Pichia pastoris. Carbohydr. Polym. 2013, 92, 784–791. [Google Scholar] [CrossRef]
- Guo, Y.; Yan, Q.; Yang, Y.; Yang, S.; Liu, Y.; Jiang, Z. Expression and characterization of a novel beta-glucosidase, with transglycosylation and exo-beta-1,3-glucanase activities, from Rhizomucor miehei. Food Chem. 2015, 175, 431–438. [Google Scholar] [CrossRef]
- Ketudat, C.J.; Esen, A. β-Glucosidases. Cell. Mol. Life Sci. CMLS 2010, 67, 3389–3405. [Google Scholar] [CrossRef]
- Hu, Y.; Luan, H.; Zhou, K.; Ge, G.; Yang, S.; Yang, L. Purification and characterization of a novel glycosidase from the china white jade snail (Achatina fulica) showing transglycosylation activity. Enzy. Microb. Technol. 2008, 43, 35–42. [Google Scholar] [CrossRef]
- Ahmed, A.; Nasim, F.H.; Batool, K.; Bibi, A. Microbial β-Glucosidase: Sources, Production and Applications. J. Appl. Environ. Microb. 2017, 5, 31–46. [Google Scholar] [CrossRef] [Green Version]
- Bourne, D.G.; Morrow, K.M.; Webster, N.S. Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annu. Rev. Microb. 2016, 70, 317–340. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Tanabe, T.; Iguchi, A. The presence of genes encoding enzymes that digest carbohydrates in coral genomes and analysis of their activities. PeerJ 2017, 5, e4087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Titlyanov, E.A.; Titlyanova, T.V.; Leletkin, V.A.; Tsukahara, J.; Van, W.R.; Yamazato, K. Degradation of zooxanthellae and regulation of their density in hermatypic corals. Mar. Ecol. Prog. Ser. 1996, 139, 167–178. [Google Scholar] [CrossRef] [Green Version]
- Krediet, C.J.; Ritchie, K.B.; Teplitski, M. Catabolite regulation of enzymatic activities in a white pox pathogen and commensal bacteria during growth on mucus polymers from the coral Acropora palmata. Dis. Aquat. Org. 2009, 87, 57–66. [Google Scholar] [CrossRef]
- Su, H.; Xiao, Z.; Yu, K.; Huang, Q.; Wang, G.; Wang, Y.; Chen, B. Diversity of cultivable protease-producing bacteria and their extracellular proteases associated to scleractinian corals. PeerJ 2020, 8, e9055. [Google Scholar] [CrossRef] [PubMed]
- Eberhart, B.; Cross, D.F.; Chase, L.R. Beta-glucosidase system of Neurospora crassa I beta-glucosidase and cellulase activities of mutant and wild-type strains. J. Bacteriol. 1964, 87, 761–770. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Mai, Z.; Yang, J.; Tian, X.; Li, J.; Zhang, S. Gene cloning and characterization of a novel salt-tolerant and glucose-enhanced beta-glucosidase from a marine Streptomycete. Appl. Biochem. Biotechnol. 2013, 169, 1512–1522. [Google Scholar] [CrossRef]
- Odoux, E.; Escoute, J.; Verdeil, J.L.; Brillouet, J.M. Localization of β-D-glucosidase activity and glucovanillin in vanilla bean (Vanilla planifolia Andrews). Annal. Botany 2003, 92, 437–444. [Google Scholar] [CrossRef] [Green Version]
- Jo, Y.Y.; Jo, K.J.; Jin, Y.L.; Kim, K.Y.; Shim, J.H.; Kim, Y.W.; Park, R.D. Characterization and kinetics of 45 kDa chitosanase from Bacillus sp. P16. Biosci. Biotechnol. Biochem. 2003, 67, 1875–1882. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Schwede, T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucl. Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Hao, S.; Liu, Y.; Qin, Y.; Zhao, L.; Zhang, J.; Wu, T.; Wang, C. Expression of a highly active β-glucosidase from Aspergillus niger AS3.4523 in Escherichia coli and its application in gardenia blue preparation. Annal. Microbiol. 2020, 70, 1–9. [Google Scholar] [CrossRef]
- Liu, L.; Zhu, S.; Zhu, T.; Zhang, M.; Wu, J.; Chen, J. Production of gentiooligosaccharide by recombinant beta-glucosidase. Wei Sheng Wu Xue Bao Acta Microbiol. Sini. 2009, 49, 597–602. [Google Scholar]
- He, H.; Qin, Y.; Chen, G.; Li, N.; Liang, Z. Two-Step purification of a novel β-glucosidase with high transglycosylation activity and another hypothetical β-glucosidase in Aspergillus oryzae HML366 and enzymatic characterization. Appl. Biochem. Biotechnol. 2013, 169, 870–884. [Google Scholar] [CrossRef] [PubMed]
- Ramani, G.; Meera, B.; Rajendhran, J.; Gunasekaran, P. Transglycosylating glycoside hydrolase family 1 β-glucosidase from Penicillium funiculosum NCL1: Heterologous expression in Escherichia coli and characterization. Biochem. Eng. J. 2015, 102, 6–13. [Google Scholar] [CrossRef]
- Seidle, H.F.; Huber, R.E. Transglucosidic reactions of the Aspergillus niger Family 3 β-glucosidase: Qualitative and quantitative analyses and evidence that the transglucosidic rate is independent of pH. Arch. Biochem. Biophys. 2005, 436, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Seidle, H.F.; Allison, S.J.; George, E.; Huber, R.E. Trp-49 of the family 3 β-glucosidase from Aspergillus niger is important for its transglucosidic activity: Creation of novel β-glucosidases with low transglucosidic efficiencies. Arch. Biochem. Biophys. 2006, 455, 110–118. [Google Scholar] [CrossRef]
| Glucose Concentration | Yields (g·L−1) | Conversion Rate (%, w/w) |
|---|---|---|
| 80% glucose | 43.02 ± 3.20 | 5.38 ± 0.40 |
| 70% glucose | 42.25 ± 4.21 | 6.07 ± 0.60 |
| 60% glucose | 40.50 ± 3.20 | 6.75 ± 1.12 |
| 50% glucose | 35.34 ± 2.60 | 7.07 ± 0.52 |
| 40% glucose | 28.21 ± 2.72 | 7.05 ± 0.68 |
| 30% glucose | 17.38 ± 0.78 | 5.79 ± 0.26 |
| 20% glucose | ND | ND |
| 10% glucose | ND | ND |
| Substrate Concentration | Yields (g·L−1) | Conversion Rate (%, w/w) |
|---|---|---|
| 60% glucose + 30% cellobiose | 51.21 ± 4.41 | 5.69 ± 0.49 |
| 50% glucose + 25% cellobiose | 56.25 ± 3.30 | 7.50 ± 0.44 |
| 40% glucose + 20% cellobiose | 63.00 ± 6.70 | 10.50 ± 1.12 |
| 30% glucose + 15% cellobiose | 70.34 ± 2.20 | 15.63 ± 0.49 |
| 25% glucose + 12.5% cellobiose | 40.39 ± 2.51 | 10.77 ± 0.66 |
| 20% glucose + 10% cellobiose | 25.38 ± 0.78 | 8.46 ± 0.26 |
| 10% glucose + 10% cellobiose | 9.02 ± 0.42 | 4.51 ± 0.21 |
| 10% glucose + 20% cellobiose | 12.84 ± 0.21 | 4.28 ± 0.07 |
| 5% glucose + 20% cellobiose | 12.88 ± 0.50 | 5.15 ± 0.20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, H.; Zhang, Q.; Yu, K.; Lu, C.; Xiao, Z.; Huang, Q.; Wang, S.; Wang, Y.; Wang, G.; Liang, J. A Novel Neutral and Mesophilic β-Glucosidase from Coral Microorganisms for Efficient Preparation of Gentiooligosaccharides. Foods 2021, 10, 2985. https://doi.org/10.3390/foods10122985
Su H, Zhang Q, Yu K, Lu C, Xiao Z, Huang Q, Wang S, Wang Y, Wang G, Liang J. A Novel Neutral and Mesophilic β-Glucosidase from Coral Microorganisms for Efficient Preparation of Gentiooligosaccharides. Foods. 2021; 10(12):2985. https://doi.org/10.3390/foods10122985
Chicago/Turabian StyleSu, Hongfei, Qi Zhang, Kefu Yu, Chunrong Lu, Zhenlun Xiao, Qinyu Huang, Shuying Wang, Yinghui Wang, Guanghua Wang, and Jiayuan Liang. 2021. "A Novel Neutral and Mesophilic β-Glucosidase from Coral Microorganisms for Efficient Preparation of Gentiooligosaccharides" Foods 10, no. 12: 2985. https://doi.org/10.3390/foods10122985
APA StyleSu, H., Zhang, Q., Yu, K., Lu, C., Xiao, Z., Huang, Q., Wang, S., Wang, Y., Wang, G., & Liang, J. (2021). A Novel Neutral and Mesophilic β-Glucosidase from Coral Microorganisms for Efficient Preparation of Gentiooligosaccharides. Foods, 10(12), 2985. https://doi.org/10.3390/foods10122985






