Comparative Peptidomics Analysis of Fermented Milk by Lactobacillus delbrueckii ssp. bulgaricus and Lactobacillus delbrueckii ssp. lactis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Culture Conditions
2.2. Fermentation of Skim Milk
2.3. Preparation of Whey Fraction
2.4. Simulated Gastrointestinal Digestion
2.5. Determination of Peptide Content
2.6. Identification of Peptides Sequence by UPLC-ESI-MS/MS
2.7. Statistical Analysis
3. Result and Discussion
3.1. Peptide Profile Analysis Revealed Variation among the Samples
3.2. Comparative Peptidomic Analysis of Fermented Milk
3.3. Comparative Peptidomic Analysis of Fermented Milk at Subspecies Level
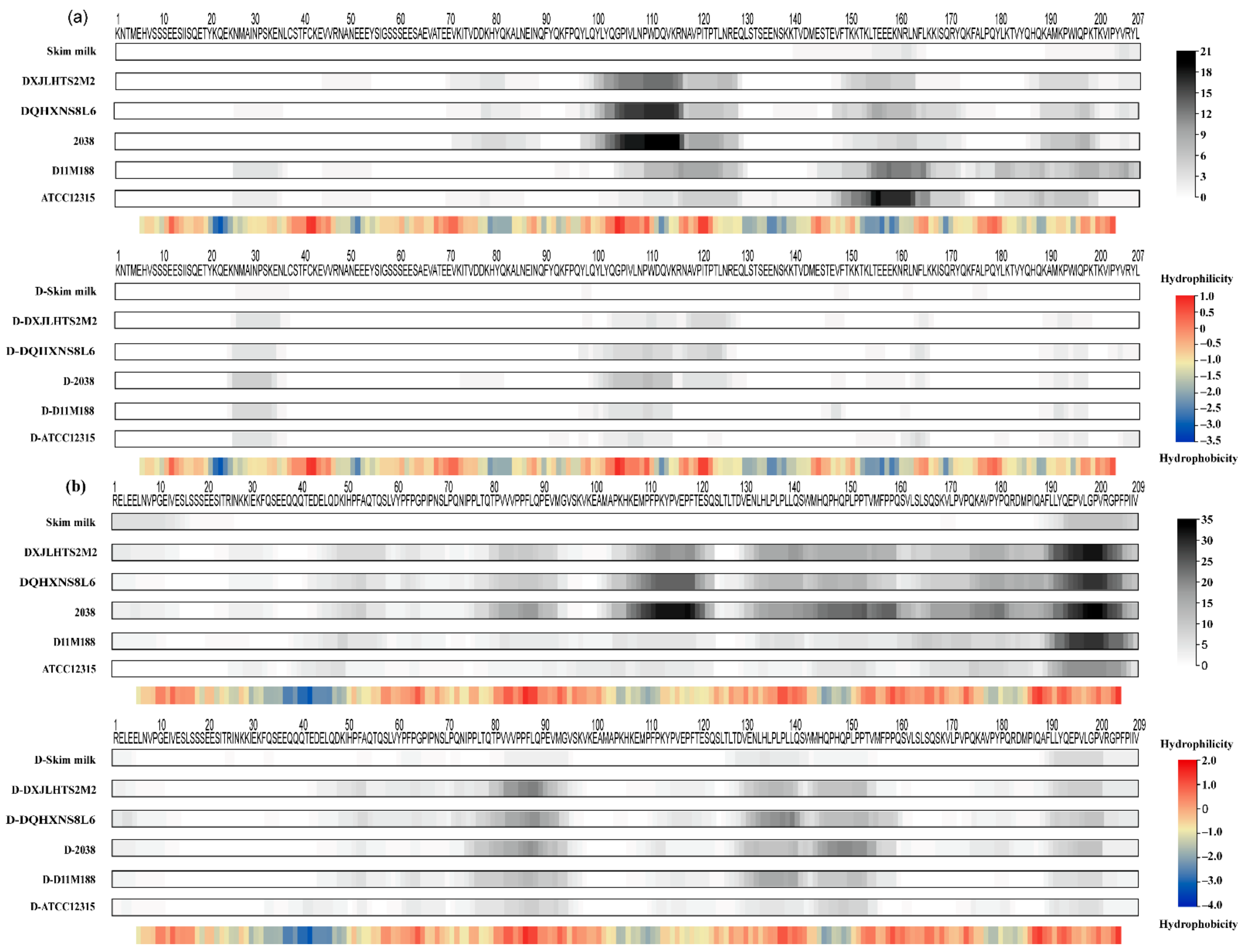
3.4. Caseins Cleavage Pattern Based on Peptidomic Analysis
3.5. Pattern of Peptidomics of Fermented Milk after Digestion
3.6. Bioactive Peptides in Fermented Milk after Digestion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weiss, N.; Schillinger, U.; Kandler, O. Lactobacillus lactis, Lactobacillus leichmannii and Lactobacillus bulgaricus, Subjective Synonyms of Lactobacillus delbrueckii, and Description of Lactobacillus delbrueckii subsp. lactis comb. nov. and Lactobacillus delbrueckii subsp. bulgaricus comb. nov. Syst. Appl. Microbiol. 1983, 4, 552–557. [Google Scholar] [CrossRef]
- El Kafsi, H.; Binesse, J.; Loux, V.; Buratti, J.; Boudebbouze, S.; Dervyn, R.; Kennedy, S.; Galleron, N.; Quinquis, B.; Batto, J.-M.; et al. Lactobacillus delbrueckii ssp. lactis and ssp. bulgaricus: A chronicle of evolution in action. BMC Genom. 2014, 15, 407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villegas, J.M.; Brown, L.; Savoy de Giori, G.; Hebert, E.M. Characterization of the mature cell surface proteinase of Lactobacillus delbrueckii subsp. lactis CRL 581. Appl. Microbiol. Biotechnol. 2015, 99, 4277–4286. [Google Scholar] [CrossRef]
- Laloi, P.; Atlan, D.; Blanc, B.; Gilbert, C.; Portalier, R. Cell-wall-associated proteinase of Lactobacillus–delbrueckii subsp. bulgaricus CNRZ 397: Differential extraction, purification and properties of the enzyme. Appl. Microbiol. Biotechnol. 1991, 36, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Tsakalidou, E.; Anastasiou, R.; Vandenberghe, I.; Van Beeumen, J.; Kalantzopoulos, G. Cell-Wall-Bound Proteinase of Lactobacillus delbrueckii subsp. lactis ACA-DC 178: Characterization and Specificity for β-Casein. Appl. Environ. Microbiol. 1999, 65, 2035–2040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, C.; Atlan, D.; Blanc, B.; Portalier, R.; Germond, J.E.; Lapierre, L.; Mollet, B. A new cell surface proteinase: Sequencing and analysis of the prtB gene from Lactobacillus delbrueckii subsp bulgaricus. J. Bacteriol. 1996, 178, 3059–3065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nongonierma, A.B.; Fitzgerald, R.J. The scientific evidence for the role of milk protein-derived bioactive peptides in humans: A Review. J. Funct. Foods 2015, 17, 640–656. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.W.; Nam, M.S. Bioactive Peptides in Milk and Dairy Products: A Review. Food Sci. Anim. Resour. 2015, 35, 831–840. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, S.D.-H.; Beverly, R.L.; Qu, Y.; Dallas, D.C. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017, 232, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Yamamoto, N.; Sakai, K.; Okubo, A.; Yamazaki, S.; Takano, T. Purification and Characterization of Angiotensin I-Converting Enzyme Inhibitors from Sour Milk. J. Dairy Sci. 1995, 78, 777–783. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yamamoto, N.; Sakai, K.; Takano, T. Antihypertensive Effect of Sour Milk and Peptides Isolated from It That are Inhibitors to Angiotensin I-Converting Enzyme. J. Dairy Sci. 1995, 78, 1253–1257. [Google Scholar] [CrossRef]
- Hata, Y.; Yamamoto, M.; Ohni, M.; Nakajima, K.; Nakamura, Y.; Takano, T. A placebo-controlled study of the effect of sour milk on blood pressure in hypertensive subjects. Am. J. Clin. Nutr. 1996, 64, 767–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohsawa, K.; Satsu, H.; Ohki, K.; Enjoh, M.; Takano, T.; Shimizu, M. Producibility and Digestibility of Antihypertensive β-Casein Tripeptides, Val-Pro-Pro and Ile-Pro-Pro, in the Gastrointestinal Tract: Analyses Using anin VitroModel of Mammalian Gastrointestinal Digestion. J. Agric. Food Chem. 2008, 56, 854–858. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, Y.; Yu, P.; Lee, Y.K.; Liu, X.; Zhao, J.; Zhang, H.; Chen, W. Effect of carbon catabolite repression on lactose and galactose catabolism in Lacticaseibacillus paracasei. Food Biosci. 2021, 40, 100912. [Google Scholar] [CrossRef]
- Wu, N.; Xu, W.; Liu, K.; Xia, Y. Shuangquan Angiotensin-converting enzyme inhibitory peptides from Lactobacillus delbrueckii QS306 fermented milk. J. Dairy Sci. 2019, 102, 5913–5921. [Google Scholar] [CrossRef] [PubMed]
- Quirós, A.; Dávalos, A.; Lasunción, M.A.; Ramos, M.; Recio, I. Bioavailability of the antihypertensive peptide LHLPLP: Transepithelial flux of HLPLP. Int. Dairy J. 2008, 18, 279–286. [Google Scholar] [CrossRef]
- Hao, X.; Yang, W.; Zhu, Q.; Zhang, G.; Zhang, X.; Liu, L.; Li, X.; Hussain, M.; Ni, C.; Jiang, X. Proteolysis and ACE-inhibitory peptide profile of Cheddar cheese: Effect of digestion treatment and different probiotics. LWT 2021, 145, 111295. [Google Scholar] [CrossRef]
- Kliche, T.; Li, B.; Bockelmann, W.; Habermann, D.; Klempt, M.; De Vrese, M.; Wutkowski, A.; Clawin-Raedecker, I.; Heller, K.J. Screening for proteolytically active lactic acid bacteria and bioactivity of peptide hydrolysates obtained with selected strains. Appl. Microbiol. Biotechnol. 2017, 101, 7621–7633. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Bayjanov, J.R.; Renckens, B.; Nauta, A.; Siezen, R.J. The proteolytic system of lactic acid bacteria revisited: A genomic comparison. BMC Genom. 2010, 11, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, D.; Ma, J.; Xu, M.; Agyei, D. Cell-envelope proteinases from lactic acid bacteria: Biochemical features and biotechnological applications. Compr. Rev. Food Sci. Food Saf. 2020, 20, 369–400. [Google Scholar] [CrossRef] [PubMed]
- Farrell, H.M., Jr.; Malin, E.L.; Brown, E.M.; Mora-Gutierrez, A. Review of the chemistry of alphaS2-casein and the generation of a homologous molecular model to explain its properties. J. Dairy Sci. 2009, 92, 1338–1353. [Google Scholar] [CrossRef] [Green Version]
- Sadat-Mekmene, L.; Jardin, J.; Corre, C.; Mollé, D.; Richoux, R.; Delage, M.-M.; Lortal, S.; Gagnaire, V. Simultaneous Presence of PrtH and PrtH2 Proteinases in Lactobacillus helveticus Strains Improves Breakdown of the Pure α s1 -Casein. Appl. Environ. Microbiol. 2011, 77, 179–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miclo, L.; Roux, E.; Genay, M.; Brusseaux, E.; Poirson, C.; Jameh, N.; Perrin, C.; Dary, A. Variability of hydrolysis of beta-, alphas1-, and alphas2-caseins by 10 strains of Streptococcus thermophilus and resulting bioactive peptides. J. Agric. Food Chem. 2012, 60, 554–565. [Google Scholar] [CrossRef]
- Holt, C.; Sawyer, L. Caseins as rheomorphic proteins: Interpretation of primary and secondary structures of the αS1-, β- and κ-caseins. J. Chem. Soc. Faraday Trans. 1993, 89, 2683–2692. [Google Scholar] [CrossRef]
- Kumosinski, T.F.; Brown, E.M.; Farrell, H.M., Jr. Three-dimensional molecular modeling of bovine caseins: An energy-minimized beta-casein structure. J. Dairy Sci. 1993, 76, 931–945. [Google Scholar] [CrossRef]
- Sabeena, F.K.H.; Baron, C.P.; Nielsen, N.S.; Otte, J.; Jacobsen, C. Antioxidant activity of yoghurt peptides: Part 2–Characterisation of peptide fractions. Food Chem. 2010, 123, 1090–1097. [Google Scholar] [CrossRef]
- Adams, C.; Sawh, F.; Green-Johnson, J.M.; Jones, T.H.; Strap, J.L. Characterization of casein-derived peptide bioactivity: Differential effects on angiotensin-converting enzyme inhibition and cytokine and nitric oxide production. J. Dairy Sci. 2020, 103, 5805–5815. [Google Scholar] [CrossRef]
- Hayes, M.; Stanton, C.; Slattery, H.; O’Sullivan, O.; Hill, C.; Fitzgerald, G.F.; Ross, R.P. Casein Fermentate of Lactobacillus animalis DPC6134 Contains a Range of Novel Propeptide Angiotensin-Converting Enzyme Inhibitors. Appl. Environ. Microbiol. 2007, 73, 4658–4667. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Rivera, L.; Diezhandino, I.; Gomez-Ruiz, J.A.; Fresno, J.M.; Miralles, B.; Recio, I. Peptidomic study of Spanish blue cheese (Valdeon) and changes after simulated gastrointestinal digestion. Electrophoresis 2014, 35, 1627–1636. [Google Scholar] [CrossRef]
- Baird, T.T.; Craik, C.S. Trypsin. In Encyclopedia of Genetics; Brenner, S., Miller, J.H., Eds.; Academic Press: New York, NY, USA, 2001; pp. 2071–2075. [Google Scholar]
- Sweeney, P.J.; Walker, J.M. Proteolytic Enzymes for Peptide Production. In Enzymes of Molecular Biology; Burrell, M.M., Ed.; Humana Press: Totowa, NJ, USA, 1993; pp. 277–303. [Google Scholar]



| Parameter | Skim Milk | DXJLHTS2M2 | DQHXNS8L6 | 2038 | D11M188 | ATCC12315 |
|---|---|---|---|---|---|---|
| Total peptides | 185 | 173 | 293 | 219 | 219 | 271 |
| Specific peptides | 83 | 24 | 77 | 34 | 29 | 87 |
| Anti-digestion peptides | 7 | 57 | 87 | 76 | 51 | 42 |
| Bioactive peptides | 16 | 22 | 37 | 26 | 35 | 28 |
| Short-sized peptides 1 | 90 | 49 | 155 | 61 | 85 | 155 |
| Medium-sized peptides 2 | 69 | 106 | 114 | 125 | 102 | 87 |
| Long-sized peptides 3 | 25 | 17 | 21 | 32 | 29 | 27 |
| Fragment | Bioactivity | Fermented Milks | D-Fermented Milks | Skim Milk | D-Skim Milk |
|---|---|---|---|---|---|
| αs1-casein f (21–22) 1 | √ | √ | √ | ||
| αs1-casein f (178–189) | √ | √ | √ | ||
| αs2-casein f (189–197)2 | √ | √ | |||
| β-casein f (78–93) | √ | √ | √ | ||
| β-casein f (130–141) | √ | √ | |||
| β-casein f (142–154) | Antioxidant | √ | √ | ||
| β-casein f (144–154) | √ | √ | √ | ||
| β-casein f (145–154) | ACE-inhibitory | √ | √ | ||
| β-casein f (192–209) | Immunomodulatory | √ | √ | √ | √ |
| κ-casein f (96–105) | √ | √ | |||
| κ-casein f (96–106) | Antioxidant | √ | √ | ||
| Fragment | Bioactivity 2 | Skim Milk | DXJLHTS2M2 | DQHXNS8L6 | 2038 | D11M188 | ATCC12315 |
|---|---|---|---|---|---|---|---|
| β-lactoglobulin f (31–32) 1 | 1 | √ | √ | √ | |||
| β-lactoglobulin f (41–60) | 10 | √ | √ | ||||
| β-lactoglobulin f (102–104) | 1, 6 | √ | √ | √ | √ | √ | √ |
| β-lactoglobulin f (125–135) | 5, 8 | √ | √ | √ | |||
| β-lactoglobulin f (104–105) | 1, 4 | √ | √ | ||||
| Lactotransferrin f (165–166) | 1 | √ | |||||
| Lactotransferrin f (166–167) | 1, 3, 4, 8 | √ | √ | ||||
| Lactotransferrin f (318–319) | 1 | √ | |||||
| serum albumin f (112–113) | 8 | √ | √ | ||||
| serum albumin f (221–222) | 1 | √ | √ | √ | √ | √ | √ |
| α-lactalbumin f (50–51) | 1 | √ | √ | ||||
| α-lactalbumin f (104–105) | 1, 8 | √ | √ | √ | √ | √ | √ |
| αs1-casein f (23–34) | 1 | √ | √ | √ | |||
| αs1-casein f (28–34) | 1 | √ | |||||
| αs1-casein f (39–40) | 6 | √ | |||||
| αs1-casein f (90–92) | 1, 6 | √ | |||||
| αs1-casein f (91–92) | 1 | √ | √ | √ | √ | ||
| αs1-casein f (91–93) | 6, 12 | √ | |||||
| αs1-casein f (91–94) | 1, 6 | √ | √ | ||||
| αs1-casein f (146–149) | 6 | √ | |||||
| αs1-casein f (165–166) | 1 | √ | |||||
| αs1-casein f (176–192) | 6 | √ | √ | ||||
| αs1-casein f (180–193) | 5 | √ | √ | √ | √ | √ | √ |
| αs1-casein f (194–199) | 1, 5 | √ | |||||
| αs1-casein f (198–199) | 1 | √ | |||||
| αs2-casein f (25–32) | 1 | √ | √ | √ | |||
| β-casein f (52–53) | 1 | √ | |||||
| β-casein f (59–68) | 1, 6 | √ | √ | ||||
| β-casein f (60–63) | 2, 14 | √ | |||||
| β-casein f (60–68) | 1, 6, 8 | √ | √ | ||||
| β-casein f (61–63) | 1 | √ | |||||
| β-casein f (61–68) | 1 | √ | |||||
| β-casein f (70–72) | 8 | √ | |||||
| β-casein f (73–89) | 1 | √ | √ | ||||
| β-casein f (78–91) | 6 | √ | √ | √ | |||
| β-casein f (84–86) | 1, 4, 6, 9, 13, 15, 16 | √ | √ | ||||
| β-casein f (106–113) | 5 | √ | √ | √ | |||
| β-casein f (130–140) | 1 | √ | √ | √ | √ | ||
| β-casein f (132–140) | 1 | √ | √ | √ | |||
| β-casein f (135–137) | 8 | √ | √ | √ | √ | ||
| β-casein f (142–154) | 6 | √ | √ | √ | √ | √ | |
| β-casein f (143–154) | 1, 4 | √ | √ | √ | √ | √ | |
| β-casein f (145–154) | 1 | √ | √ | √ | √ | √ | |
| β-casein f (145–160) | 1, 4 | √ | √ | √ | |||
| β-casein f (151–153) | 1 | √ | |||||
| β-casein f (169–176) | 6 | √ | |||||
| β-casein f (191–193) | 4, 6, 11 | √ | √ | √ | |||
| β-casein f (191–202) | 1 | √ | √ | √ | √ | √ | √ |
| β-casein f (191–209) | 1 | √ | |||||
| β-casein f (192–202) | 1, 4 | √ | √ | √ | √ | √ | √ |
| β-casein f (192–209) | 11 | √ | √ | √ | √ | √ | √ |
| β-casein f (193–202) | 1, 4, 6, 7, 11 | √ | √ | √ | √ | √ | |
| β-casein f (193–209) | 1, 5, 7, 11 | √ | √ | √ | √ | √ | √ |
| κ-casein f (7–8) | 1 | √ | |||||
| κ-casein f (14–17) | 1 | √ | √ | √ | |||
| κ-casein f (30–32) | 5, 6 | √ | √ | √ | |||
| κ-casein f (58–60) | 8 | √ | |||||
| κ-casein f (96–106) | 6 | √ | √ | √ | √ | √ | |
| κ-casein f (97–106) | 6 | √ | √ | √ | √ | ||
| κ-casein f (150–151) 3 | 1 | √ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, H.; Zhang, X.; Jiang, Y.; Guo, M.; Liu, X.; Zhao, J.; Zhang, H.; Chen, W. Comparative Peptidomics Analysis of Fermented Milk by Lactobacillus delbrueckii ssp. bulgaricus and Lactobacillus delbrueckii ssp. lactis. Foods 2021, 10, 3028. https://doi.org/10.3390/foods10123028
Ye H, Zhang X, Jiang Y, Guo M, Liu X, Zhao J, Zhang H, Chen W. Comparative Peptidomics Analysis of Fermented Milk by Lactobacillus delbrueckii ssp. bulgaricus and Lactobacillus delbrueckii ssp. lactis. Foods. 2021; 10(12):3028. https://doi.org/10.3390/foods10123028
Chicago/Turabian StyleYe, Hongji, Xinyi Zhang, Yang Jiang, Min Guo, Xiaoming Liu, Jianxin Zhao, Hao Zhang, and Wei Chen. 2021. "Comparative Peptidomics Analysis of Fermented Milk by Lactobacillus delbrueckii ssp. bulgaricus and Lactobacillus delbrueckii ssp. lactis" Foods 10, no. 12: 3028. https://doi.org/10.3390/foods10123028






