Comparative Study on Pale, Soft and Exudative (PSE) and Red, Firm and Non-Exudative (RFN) Pork: Protein Changes during Aging and the Differential Protein Expression of the Myofibrillar Fraction at 1 h Postmortem
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. The pH, Color, and Purge Loss
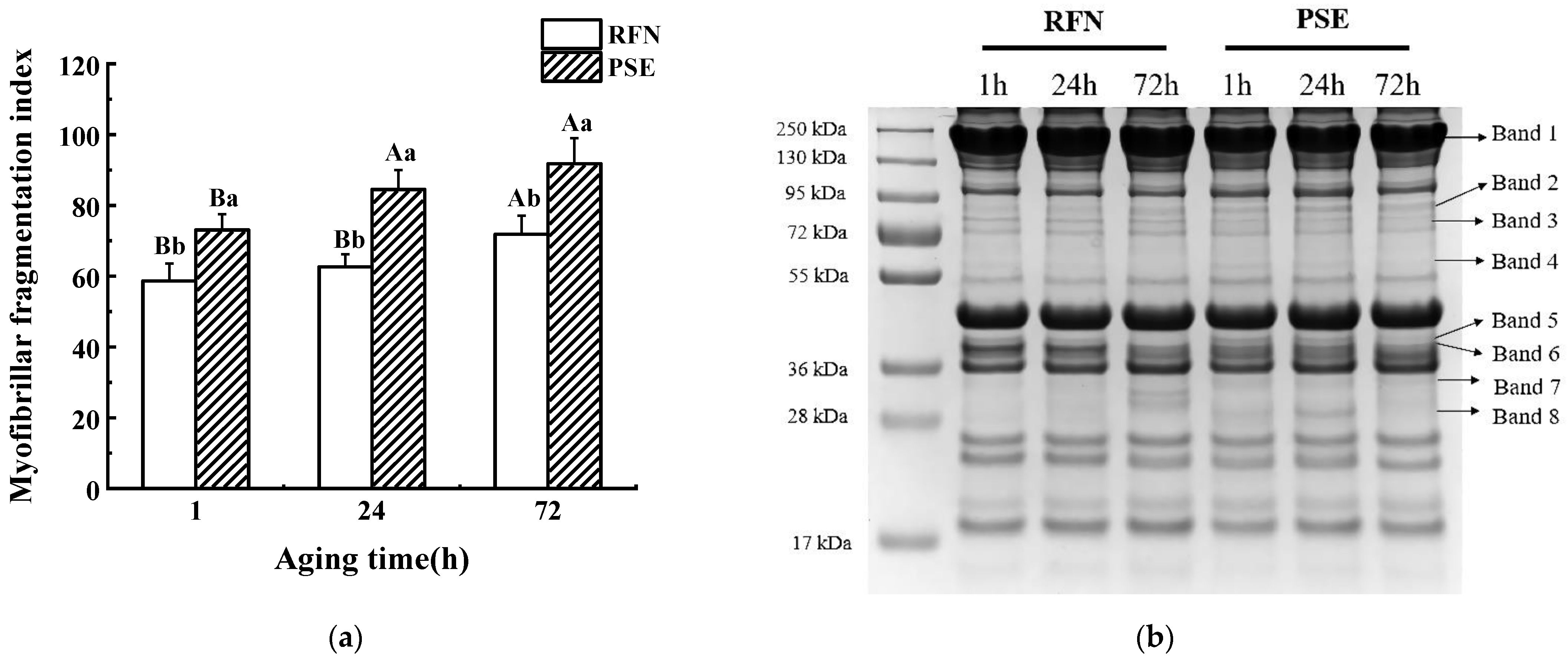
2.3. The Myofibrillar Fragmentation Index
2.4. SDS-PAGE and Western Blotting
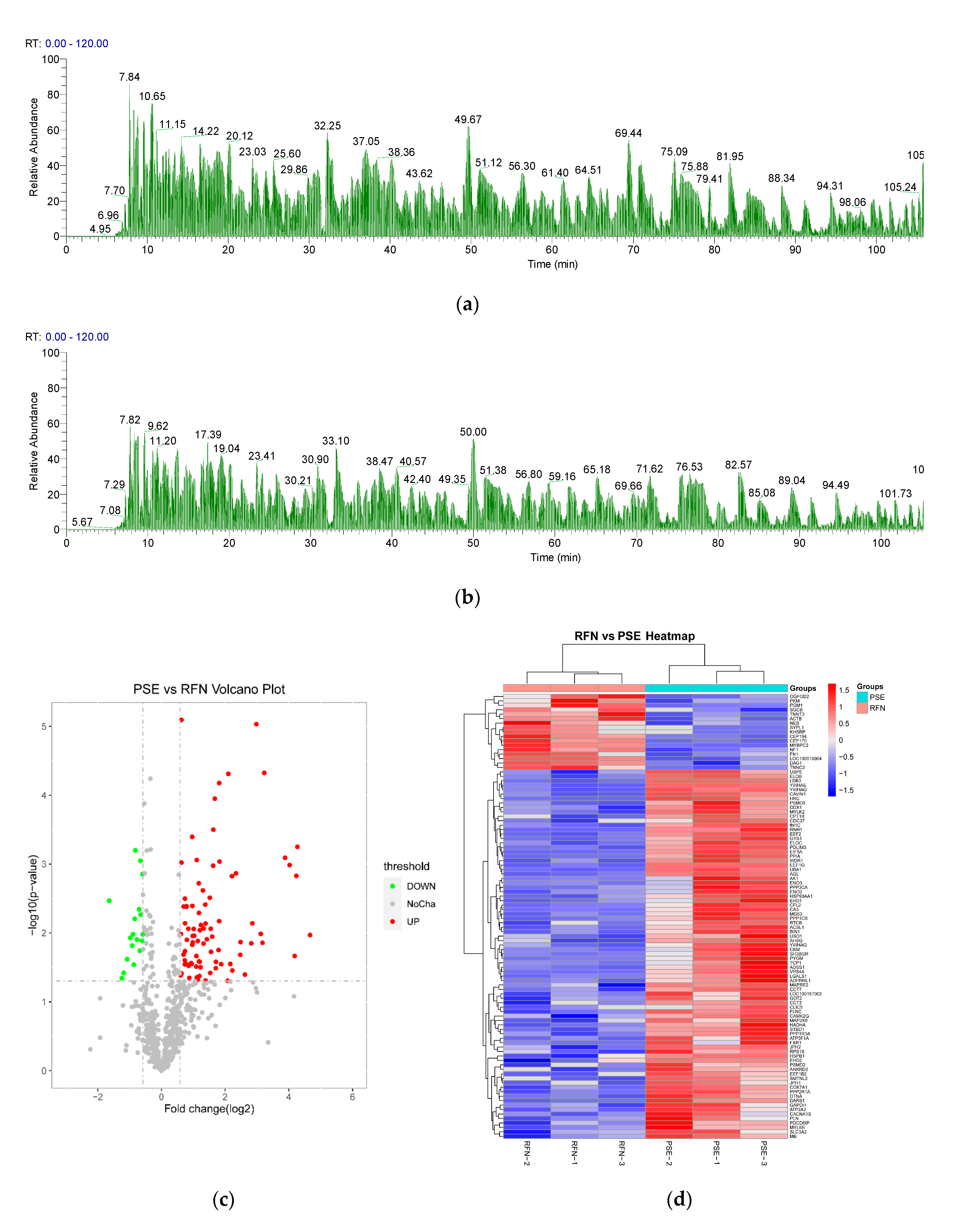
2.5. LC-MS/MS Analysis
2.6. Bioinformatics Analysis
2.7. Statistical Analysis
3. Results
3.1. Purge Loss, pH, and Color
3.2. MFI and SDS-PAGE
3.3. Desmin Degradation
3.4. Myofibrillar Protein Identification and Quantification
3.5. GO Functional Annotation of the Differential Proteins
3.6. KEGG Pathway Analysis and PPI Network of the Differential Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Trevisan, L.; Brum, J.S. Incidence of pale, soft and exudative (PSE) pork meat in reason of extrinsic stress factors. Anais Acad. Bras. Ciências 2020, 92, e20190086. [Google Scholar] [CrossRef]
- Karamucki, T.; Jakubowska, M.; Rybarczyk, A.; Gardzielewska, J. The influence of myoglobin on the colour of minced pork loin. Meat Sci. 2013, 94, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, H.; Qi, J.; Wang, M.; Xu, X.; Zhou, G. Chicken breast quality—Normal, pale, soft and exudative (PSE) and woody—Influences the functional properties of meat batters. Int. J. Food Sci. Technol. 2018, 53, 654–664. [Google Scholar] [CrossRef]
- Li, X.; Feng, F.; Gao, R.; Wang, L.; Qian, Y.; Li, C.; Zhou, G. Application of near infrared reflectance (NIR) spectroscopy to identify potential PSE meat. J. Sci. Food Agric. 2015, 96, 3148–3156. [Google Scholar] [CrossRef]
- Adzitey, F.; Nurul, H. Pale soft exudative (PSE) and dark firm dry (DFD) meats: Causes and measures to reduce these incidences-a mini review. Int. Food Res. J. 2011, 18, 11–20. [Google Scholar]
- Choi, Y.; Kim, B. Muscle fiber characteristics, myofibrillar protein isoforms, and meat quality. Livest. Sci. 2009, 122, 105–118. [Google Scholar] [CrossRef]
- Zequan, X.; Yonggang, S.; Guangjuan, L.; Shijun, X.; Li, Z.; Mingrui, Z.; Yanli, X.; Zirong, W. Proteomics analysis as an approach to understand the formation of pale, soft, and exudative (PSE) pork. Meat Sci. 2021, 177, 108353. [Google Scholar] [CrossRef]
- Goll, D.E.; Neti, G.; Mares, S.W.; Thompson, V.F. Myofibrillar protein turnover: The proteasome and the calpains1,2. J. Anim. Sci. 2008, 86, E19–E35. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Hou, C.; Ijaz, M.; Yan, T.; Li, X.; Li, Y.; Zhang, D. Proteomics discovery of protein biomarkers linked to meat quality traits in post-mortem muscles: Current trends and future prospects: A review. Trends Food Sci. Technol. 2020, 105, 416–432. [Google Scholar] [CrossRef]
- Carlson, K.B.; Prusa, K.J.; Fedler, C.A.; Steadham, E.M.; Outhouse, A.C.; King, D.A.; Huff-Lonergan, E.; Lonergan, S.M. Postmortem protein degradation is a key contributor to fresh pork loin tenderness123. J. Anim. Sci. 2017, 95, 1574–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Zhou, G.-H.; Zhang, W.-G. Effects of High Oxygen Packaging on Tenderness and Water Holding Capacity of Pork Through Protein Oxidation. Food Bioprocess. Technol. 2015, 8, 2287–2297. [Google Scholar] [CrossRef]
- Wojtysiak, D.; Górska, M. Effect of Aging Time on Meat Quality and Rate of Desmin and Dystrophin Degradation of Pale, Soft, Exudative (PSE) and Normal Turkey Breast Muscle. Folia Biol. 2018, 66, 63–72. [Google Scholar] [CrossRef]
- Yin, Y.; Zhang, W.G.; Zhou, G.H.; Guo, B. Comparison of protein degradation, protein oxidation, and μ-calpain activation between pale, soft, and exudative and red, firm, and nonexudative pork during postmortem aging1. J. Anim. Sci. 2014, 92, 3745–3752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van De Wiel, D.F.; Zhang, W.L. Identification of pork quality parameters by proteomics. Meat Sci. 2007, 77, 46–54. [Google Scholar] [CrossRef]
- Hwang, I.; Park, B.; Cho, S.; Lee, J. Effects of muscle shortening and proteolysis on Warner–Bratzler shear force in beef longissimus and semitendinosus. Meat Sci. 2004, 68, 497–505. [Google Scholar] [CrossRef]
- Pearce, K.L.; Rosenvold, K.; Andersen, H.J.; Hopkins, D.L. Water distribution and mobility in meat during the conversion of muscle to meat and ageing and the impacts on fresh meat quality attributes—A review. Meat Sci. 2011, 89, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Chmiel, M.; Słowiński, M.; Janakowski, S. The Quality Evaluation of RFN and PSE Pork Longissimus Lumborum Muscle Considering Its Microstructure. Ann. Anim. Sci. 2014, 14, 737–747. [Google Scholar] [CrossRef] [Green Version]
- Warner, R.; Kauffman, R.; Greaser, M. Muscle protein changes post mortem in relation to pork quality traits. Meat Sci. 1997, 45, 339–352. [Google Scholar] [CrossRef]
- Lomiwes, D.; Farouk, M.; Wu, G.; Young, O. The development of meat tenderness is likely to be compartmentalised by ultimate pH. Meat Sci. 2014, 96, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, S.; Yan, P. The meat quality, muscle fiber characteristics and fatty acid profile in Jinjiang and F1 Simmental×Jinjiang yellow cattle. Asian-Australas. J. Anim. Sci. 2018, 31, 301–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardoso, G.P.; Dutra, M.P.; Fontes, P.R.; Ramos, A.D.L.S.; Gomide, L.A.D.M.; Ramos, E.M. Selection of a chitosan gelatin-based edible coating for color preservation of beef in retail display. Meat Sci. 2016, 114, 85–94. [Google Scholar] [CrossRef]
- Qian, S.; Li, X.; Wang, H.; Wei, X.; Mehmood, W.; Zhang, C.; Blecker, C. Contribution of calpain to protein degradation, variation in myowater properties and the water-holding capacity of pork during postmortem ageing. Food Chem. 2020, 324, 126892. [Google Scholar] [CrossRef] [PubMed]
- Gornall, A.G.; Bardawill, C.J.; David, M.M. Determination of serum proteins by means of the biuret reaction. J. Biol. Chem. 1949, 177, 751–766. [Google Scholar] [CrossRef]
- Xiong, Y.; Brekke, C. Gelation properties of chicken myofibrils treated with calcium and magnesium chlorides. J. Muscle Foods 1991, 2, 21–36. [Google Scholar] [CrossRef]
- Zhang, W.; Lonergan, S.M.; Gardner, M.A.; Huff-Lonergan, E. Contribution of postmortem changes of integrin, desmin and μ-calpain to variation in water holding capacity of pork. Meat Sci. 2006, 74, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Huan, T. Comparison of Full-Scan, Data-Dependent, and Data-Independent Acquisition Modes in Liquid Chromatography–Mass Spectrometry Based Untargeted Metabolomics. Anal. Chem. 2020, 92, 8072–8080. [Google Scholar] [CrossRef]
- Fernández-Costa, C.; Martínez-Bartolomé, S.; McClatchy, D.B.; Saviola, A.J.; Yu, N.-K.; Yates, I.J.R. Impact of the Identification Strategy on the Reproducibility of the DDA and DIA Results. J. Proteome Res. 2020, 19, 3153–3161. [Google Scholar] [CrossRef] [PubMed]
- Swatland, H. How pH causes paleness or darkness in chicken breast meat. Meat Sci. 2008, 80, 396–400. [Google Scholar] [CrossRef]
- Guo, B.; Zhang, W.; Tume, R.K.; Hudson, N.J.; Huang, F.; Yin, Y.; Zhou, G. Disorder of endoplasmic reticulum calcium channel components is associated with the increased apoptotic potential in pale, soft, exudative pork. Meat Sci. 2016, 115, 34–40. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, R.; Tian, X.; Fan, X.; Shi, Y.; Zhang, W.; Hou, Q.; Zhou, G. Comparison of Activity, Expression, and S-Nitrosylation of Calcium Transfer Proteins between Pale, Soft, and Exudative and Red, Firm, and Non-exudative Pork during Post-Mortem Aging. J. Agric. Food Chem. 2019, 67, 3242–3248. [Google Scholar] [CrossRef] [PubMed]
- Gokhin, D.S.; Fowler, V.M. A two-segment model for thin filament architecture in skeletal muscle. Nat. Rev. Mol. Cell Biol. 2013, 14, 113–119. [Google Scholar] [CrossRef] [Green Version]
- Veiseth, E.; Shackelford, S.D.; Wheeler, T.L.; Koohmaraie, M. Technical note: Comparison of myofibril fragmentation index from fresh and frozen pork and lamb longissimus. J. Anim. Sci. 2001, 79, 904–906. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, A.E.; Maganhini, M.B.; Hernández-Blazquez, F.J.; Ida, E.I.; Shimokomaki, M. Protease activity and the ultrastructure of broiler chicken PSE (pale, soft, exudative) meat. Food Chem. 2010, 119, 1201–1204. [Google Scholar] [CrossRef]
- Goldmann, W.H. Intermediate filaments and cellular mechanics. Cell Biol. Int. 2018, 42, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Jia, S.; Liu, J.; Gao, P.; Yu, D.; Jiang, Q.; Xu, Y.; Yu, P.; Xia, W.; Zhan, X. The relationship between degradation of myofibrillar structural proteins and texture of superchilled grass carp (Ctenopharyngodon idella) fillet. Food Chem. 2019, 301, 125278. [Google Scholar] [CrossRef]
- Liu, J.; Arner, A.; Puolanne, E.; Ertbjerg, P. On the water-holding of myofibrils: Effect of sarcoplasmic protein denaturation. Meat Sci. 2016, 119, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Ertbjerg, P.; Puolanne, E. Muscle structure, sarcomere length and influences on meat quality: A review. Meat Sci. 2017, 132, 139–152. [Google Scholar] [CrossRef] [Green Version]
- Kremneva, E.; Makkonen, M.H.; Skwarek-Maruszewska, A.; Gateva, G.; Michelot, A.; Dominguez, R.; Lappalainen, P. Cofilin-2 Controls Actin Filament Length in Muscle Sarcomeres. Dev. Cell 2014, 31, 215–226. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Takeda, S.; Mitsuoka, K.; Oda, T.; Kimura-Sakiyama, C.; Maéda, Y.; Narita, A. Structural basis for cofilin binding and actin filament disassembly. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Watanabe, A.; Devine, C. Effect of meat ultimate pH on rate of titin and nebulin degradation. Meat Sci. 1996, 42, 407–413. [Google Scholar] [CrossRef]
- Tskhovrebova, L.; Trinick, J. Titin and Nebulin in Thick and Thin Filament Length Regulation. Subcell. Biochem. 2017, 82, 285–318. [Google Scholar] [CrossRef]
- Hwang, I.; Park, B.; Kim, J.; Cho, S.; Lee, J. Assessment of postmortem proteolysis by gel-based proteome analysis and its relationship to meat quality traits in pig longissimus. Meat Sci. 2005, 69, 79–91. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, D.; Geng, Z.; Bian, H.; Liu, F.; Zhu, Y.; Xu, W. The level of heat shock protein 90 in pig Longissimus dorsi muscle and its relationship with meat pH and quality. Food Chem. 2014, 165, 337–341. [Google Scholar] [CrossRef]
- Paulsen, G.; Vissing, K.; Kalhovde, J.M.; Ugelstad, I.; Bayer, M.L.; Kadi, F.; Schjerling, P.; Hallén, J.; Raastad, T. Maximal eccentric exercise induces a rapid accumulation of small heat shock proteins on myofibrils and a delayed HSP70 response in humans. Am. J. Physiol. Integr. Comp. Physiol. 2007, 293, R844–R853. [Google Scholar] [CrossRef] [Green Version]
- Sandercock, D.A.; Hunter, R.R.; Nute, G.R.; Mitchell, M.A.; Hocking, P.M. Acute Heat Stress-Induced Alterations in Blood Acid-Base Status and Skeletal Muscle Membrane Integrity in Broiler Chickens at Two Ages: Implications for Meat Quality. Poult. Sci. 2001, 80, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, M.; Bian, H.; Xu, W.; Xu, X.; Zhu, Y.; Liu, F.; Geng, Z.; Zhou, G. Changes of phospholipase A2 and C activities during dry-cured duck processing and their relationship with intramuscular phospholipid degradation. Food Chem. 2014, 145, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Carra, S.; Alberti, S.; Benesch, J.L.P.; Boelens, W.; Buchner, J.; Carver, J.A.; Cecconi, C.; Ecroyd, H.; Gusev, N.; Hightower, L.E.; et al. Small heat shock proteins: Multifaceted proteins with important implications for life. Cell Stress Chaperon 2019, 24, 295–308. [Google Scholar] [CrossRef]
- Doberentz, E.; Genneper, L.; Wagner, R.; Madea, B. Expression times for hsp27 and hsp70 as an indicator of thermal stress during death due to fire. Int. J. Leg. Med. 2017, 131, 1707–1718. [Google Scholar] [CrossRef]
- Ramanathan, R.; Suman, S.P.; Faustman, C. Biomolecular Interactions Governing Fresh Meat Color in Post-mortem Skeletal Muscle: A Review. J. Agric. Food Chem. 2020, 68, 12779–12787. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.G.; Brewer, M.S. Effects of urea denatureation and pH on the ability of porcine myoglobin to undergo reduction. Meat Sci. 2003, 63, 427–432. [Google Scholar] [CrossRef]
- Lambert, C.M.; Roy, M.; Robitaille, G.A.; Richard, D.E.; Bonnet, S. HIF-1 inhibition decreases systemic vascular remodelling diseases by promoting apoptosis through a hexokinase 2-dependent mechanism. Cardiovasc. Res. 2010, 88, 196–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surazynski, A.; Donald, S.P.; Cooper, S.K.; Whiteside, M.A.; Salnikow, K.; Liu, Y.; Phang, J.M. Extracellular matrix and HIF-1 signaling: The role of prolidase. Int. J. Cancer 2008, 122, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Nguyen, P.T.; Wang, X.; Zhao, Y.; Meacham, C.E.; Zou, Z.; Bordieanu, B.; Johanns, M.; Vertommen, D.; Wijshake, T.; et al. TLR9 and beclin 1 crosstalk regulates muscle AMPK activation in exercise. Nat. Cell Biol. 2020, 578, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Ke, R.; Xu, Q.; Li, C.; Luo, L.; Huang, D. Mechanisms of AMPK in the maintenance of ATP balance during energy metabolism. Cell Biol. Int. 2018, 42, 384–392. [Google Scholar] [CrossRef] [Green Version]
- Kuo, I.Y.; Ehrlich, B.E. Signaling in Muscle Contraction. Cold Spring Harb. Perspect. Biol. 2015, 7, a006023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yao, L.; Yang, J.; Wang, Z.; Du, G. PI3K/Akt and HIF-1 signaling pathway in hypoxia-ischemia (Review). Mol. Med. Rep. 2018, 18, 3547–3554. [Google Scholar] [CrossRef] [Green Version]



| Index | Postmortem Time(h) | RFN | PSE |
|---|---|---|---|
| pH | 1 | 5.88 ± 0.33 a | 5.48 ± 0.16 b |
| 24 | 5.63 ± 0.26 a | 5.51 ± 0.29 b | |
| L* | 1 | 42.64 ± 0.93 b | 53.27 ± 2.24 a |
| 24 | 46.85 ± 0.67 b | 56.06 ± 2.12 a | |
| Purge loss(%) | 24 | 1.60 ± 0.44 b | 6.96 ± 1.99 a |
| 72 | 2.84 ± 0.72 b | 8.12 ± 3.46 a |
| RFN | PSE | |||||
|---|---|---|---|---|---|---|
| 1 | 24 | 72 | 1 | 24 | 72 | |
| Band 1 | 1.08 ± 0.01 C,a | 1.13 ± 0.03 B,a | 1.42 ± 0.02 A,a | 1.00 ± 0.00 B,b | 1.04 ± 0.01 A,b | 1.03 ± 0.01 A,b |
| Band 2 | 0.92 ± 0.01 C,a | 0.95 ± 0.01 B,a | 1.00 ± 0.02 A,a | 1.00 ± 0.00 B,a | 1.08 ± 0.01 A,a | 0.96 ± 0.01 C,a |
| Band 3 | ND | ND | ND | 1.00 ± 0.00 A | 0.99 ± 0.01 B | 0.91 ± 0.01 C |
| Band 4 | 0.91 ± 0.01 A,b | 0.96 ± 0.01 A,b | 0.91 ± 0.01 A,a | 1.00 ± 0.00 A,a | 0.98 ± 0.01 B,a | 0.87 ± 0.01 C,b |
| Band 5 | 0.99 ± 0.04 AB,a | 1.19 ± 0.15 A,a | 0.79 ± 0.01 B,a | 1.00 ± 0.00 A,a | 1.04 ± 0.03 A,a | 0.89 ± 0.03 B,a |
| Band 6 | 2.37 ± 0.08 A,a | 2.57 ± 0.23 A,a | 0.88 ± 0.04 B,a | 0.99 ± 0.00 A,b | 1.02 ± 0.08 A,b | 0.95 ± 0.04 A,a |
| Band 7 | 0.85 ± 0.01 C,b | 0.93 ± 0.01 B,b | 0.98 ± 0.01 A,a | 1.00 ± 0.00 A,a | 0.97 ± 0.01 B,a | 0.96 ± 0.01 B,b |
| Band 8 | ND | ND | ND | 1.00 ± 0.00 B | 1.07 ± 0.02 A | 0.85 ± 0.01 C |
| No. | Protein Names | Protein UniProt/ NCBI Accession | Gene Names | Score | FC | p-Value |
|---|---|---|---|---|---|---|
| Microfilaments | ||||||
| 1 | Beta actin (Fragment) | Q00P29 | ACTB | 3.97 | 0.64 | <0.01 |
| 2 | Cofilin-2 | Q5G6V9 | CFL2 | 40.98 | 9.02 | <0.05 |
| 3 | Nebulin | A0A287B9W0 | NEB | 17.18 | 0.44 | <0.05 |
| Microtubules | ||||||
| 1 | Microtubule-associated protein RP/EB family member 2 | A0A4X1VCG5 | MAPRE2 | 15.03 | 1.74 | <0.05 |
| Muscle contraction | ||||||
| 1 | Troponin T | A0A4X1U8T2 | TNNT3 | 323.31 | 0.59 | <0.05 |
| 2 | Voltage-dependent L-type calcium channel subunit alpha | A0A5G2QSL4 | CACNA1S | 43.95 | 1.55 | <0.05 |
| 3 | Cardiac phospholamban | P61013 | PLN | 3.11 | 3.26 | <0.05 |
| Motor proteins | ||||||
| 1 | Myosin heavy chain (Fragment) | Q95249 | / | 6.87 | 0.54 | <0.05 |
| 2 | Dynein light chain | D9U8D1 | DYNLL1 | 16.91 | * | * |
| Other sarcomeric proteins | ||||||
| 1 | PDZ and LIM domain protein 3 | Q6QGC0 | PDLIM3 | 305.84 | 3.08 | <0.01 |
| 2 | Beta-sarcoglycan | F1SE70 | SGCB | 20.01 | 0.55 | <0.05 |
| 3 | IF rod domain-containing protein | A0A4X1UHN0 | / | 29.05 | 2.13 | <0.05 |
| 4 | WD repeat-containing protein 1 | K9IVR7 | WDR1 | 177.78 | 5.06 | <0.01 |
| Glycogen metabolism | ||||||
| 1 | Glycogen synthase | A0A5G2QKR0 | GYS1 | 67.92 | 2.46 | <0.01 |
| 2 | 4-alpha-glucanotransferase | F1S557 | AGL | 128.07 | 4.29 | <0.01 |
| Glycolysis | ||||||
| 1 | Pyruvate kinase | A0A480JGH8 | PKM | 49.58 | 0.42 | <0.05 |
| 2 | 2-phospho-D-glycerate hydro-lyase | A0A4X1UZ92 | ENO2 | 5.12 | 2.29 | <0.05 |
| 3 | Glyceraldehyde-3-phosphate dehydrogenase | P00355 | GAPDH | 214.00 | 1.72 | <0.05 |
| 4 | Creatine kinase | A0A5G2QZN6 | CKM | 323.31 | 4.68 | <0.05 |
| Energy metabolism | ||||||
| 1 | Cytochrome c oxidase subunit 7A1 | Q8SPJ9 | COX7A1 | 3.38 | 1.67 | <0.01 |
| 2 | Calcium-transporting ATPase | A0A480TDT7 | ATP2A2 | 295.12 | 2.50 | <0.05 |
| 3 | Adenylosuccinate synthetase isozyme 1 | A0A287BAF3 | ADSS1 | 105.07 | 7.07 | <0.05 |
| Transport | ||||||
| 1 | Eukaryotic translation initiation factor 5A | A0A4X1V2D0 | EIF5A | 98.96 | 19.27 | <0.01 |
| Molecular chaperones | ||||||
| 1 | Heat shock protein HSP 90-alpha isoform 2 | A0A481CXT9 | HSP90AA1 | 256.08 | 3.02 | <0.05 |
| 2 | Heat shock protein beta-1 | Q5S1U1 | HSPB1 | 323.31 | 1.67 | <0.05 |
| 3 | Hsp90 chaperone protein kinase-targeting subunit | F6Q4F9 | CDC37 | 10.90 | 2.26 | <0.05 |
| 4 | T-complex protein 1 subunit | A0A5G2RH19 | CCT7 | 11.29 | 1.62 | <0.01 |
| 5 | T-complex protein 1 subunit gamma | A0A480WDC3 | CCT3 | 27.42 | 2.31 | <0.01 |
| Enzyme proteins | ||||||
| 1 | Dual specificity protein phosphatase | I3LCX3 | DUSP3 | 13.52 | * | * |
| 2 | 26S proteasome -ATPase subunit RPT1 | A0A480VIW9 | PSMC2 | 11.63 | * | * |
| 3 | AMPKBI domain-containing protein | K7GPQ2 | PRKAB2 | 3.15 | * | * |
| 4 | Flavin reductase (NADPH) | I3LQH7 | BLVRB | 3.95 | * | * |
| 5 | Phosphoinositide phospholipase C | F1SRY6 | PLCD4 | 28.16 | * | * |
| 6 | 5-AMP-activated protein kinase subunit gamma-3 | K7GM96 | PRKAG3 | 2.49 | * | * |
| 7 | Peptidyl-prolyl cis-trans isomerase | I3LLH5 | PIN1 | 20.52 | * | * |
| 8 | S-formylglutathione hydrolase | A0A5S6IDI6 | ESD | 3.83 | * | * |
| 9 | Ubiquitin carboxyl-terminal hydrolase | A0A4X1VR78 | USP14 | 7.39 | * | * |
| 10 | Cathepsin B | A0A287BF94 | CTSB | 7.84 | ** | ** |
| 11 | Ubiquitin-activating enzyme E1 | A0A480W380 | UBA1 | 51.87 | 9.38 | <0.01 |
| 12 | Calcium/calmodulin-dependent protein kinase | F6QB46 | CAMK2G | 49.06 | 1.66 | <0.05 |
| 13 | Alpha-1,4 glucan phosphorylase | A0A286ZMZ9 | PYGM | 323.31 | 18.18 | <0.05 |
| 14 | Adenylate kinase isoenzyme 1 | P00571 | AK1 | 187.73 | 6.16 | <0.05 |
| 15 | ATP synthase subunit alpha | F1RPS8 | ATP5F1A | 63.03 | 1.97 | <0.05 |
| 16 | Aspartate aminotransferase | A0A4X1UT32 | GOT2 | 17.00 | 1.62 | <0.05 |
| 17 | Striated muscle preferentially expressed protein kinase | A0A481BRQ9 | / | 64.82 | 1.53 | <0.05 |
| 18 | 26S proteasome non-ATPase regulatory subunit 2 | I3LEW5 | PSMD2 | 14.26 | 2.89 | <0.05 |
| 19 | Aspartate-tRNA ligase, cytoplasmic | A0A480NFZ5 | DARS1 | 29.47 | 1.54 | <0.01 |
| 20 | Myosin light chain kinase 2 | F1S7H3 | MYLK2 | 177.41 | 1.96 | <0.01 |
| 21 | Vesicle-fusing ATPase | A0A4X1SF26 | VPS4A | 16.74 | 2.02 | <0.01 |
| 22 | Protein kinase domain-containing protein | A0A5K1UKA4 | MAP2K6 | 23.01 | 1.91 | <0.05 |
| 23 | Serine/threonine-protein phosphatase | A0A286ZXJ6 | PPP3CA | 7.29 | 2.61 | <0.05 |
| 24 | Serine/threonine-protein phosphatase PP1-beta catalytic subunit | P61292 | PPP1CB | 26.24 | 7.24 | <0.01 |
| 25 | Ubiquitin carboxyl-terminal hydrolase | A0A5G2R3H5 | USP5 | 27.48 | 2.61 | <0.01 |
| Others | ||||||
| 1 | Peptidyl-prolyl cis-trans isomerase A | P62936 | PPIA | 12.36 | 18.84 | <0.01 |
| 2 | Myoglobin | P02189 | MB | 97.32 | 2.21 | <0.05 |
| 3 | Alpha-dystroglycan | A0A286ZY59 | DAG1 | 26.12 | 0.56 | <0.01 |
| 4 | Carbonic anhydrase 3 | Q5S1S4 | CA3 | 323.31 | 25.40 | <0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Wu, G.-Y.; Li, K.-Y.; Ge, Q.-F.; Wu, M.-G.; Yu, H.; Wu, S.-L.; Bao, W.-B. Comparative Study on Pale, Soft and Exudative (PSE) and Red, Firm and Non-Exudative (RFN) Pork: Protein Changes during Aging and the Differential Protein Expression of the Myofibrillar Fraction at 1 h Postmortem. Foods 2021, 10, 733. https://doi.org/10.3390/foods10040733
Liu R, Wu G-Y, Li K-Y, Ge Q-F, Wu M-G, Yu H, Wu S-L, Bao W-B. Comparative Study on Pale, Soft and Exudative (PSE) and Red, Firm and Non-Exudative (RFN) Pork: Protein Changes during Aging and the Differential Protein Expression of the Myofibrillar Fraction at 1 h Postmortem. Foods. 2021; 10(4):733. https://doi.org/10.3390/foods10040733
Chicago/Turabian StyleLiu, Rui, Guo-Yue Wu, Ke-Yue Li, Qing-Feng Ge, Man-Gang Wu, Hai Yu, Sheng-Long Wu, and Wen-Bin Bao. 2021. "Comparative Study on Pale, Soft and Exudative (PSE) and Red, Firm and Non-Exudative (RFN) Pork: Protein Changes during Aging and the Differential Protein Expression of the Myofibrillar Fraction at 1 h Postmortem" Foods 10, no. 4: 733. https://doi.org/10.3390/foods10040733








