Lactic Acid Bacteria Diversity and Characterization of Probiotic Candidates in Fermented Meats
Abstract
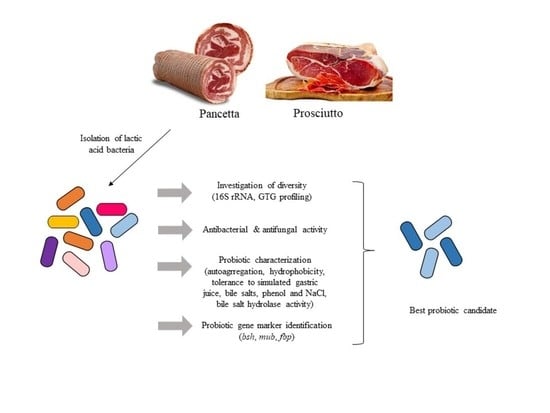
:1. Introduction
2. Materials and Methods
2.1. LAB Diversity, Organic Acid Profiling and Antibiotic Susceptibility
2.1.1. Isolation and Growth Condition
2.1.2. Species Identification Using 16S rRNA Sequence Analysis
2.1.3. Phylogenetic Inference
2.1.4. Genetic Fingerprinting by (GTG)5 PCR
2.1.5. Identification and Quantification of Organic Acid Production
2.1.6. Antibiotic Susceptibility
2.2. Antagonistic Activity
2.2.1. Bacteria Indicator Strains
2.2.2. Spot on Lawn Antibacterial Assay
2.2.3. Well Diffusion Assay to Detect Bacteriocin Production
2.2.4. Spot-on-Lawn Antifungal Assay
2.3. Screening for Probiotic Candidates
2.3.1. Phenol and Salt Tolerance
2.3.2. Bile Salt Tolerance
2.3.3. Simulated Gastric Juice Tolerance
2.3.4. Analysis of Autoaggregation and Hydrophobicity Properties
2.4. Bioinformatic Identification of Bacteriocin Gene Clusters and Probiotic Gene Markers
2.4.1. DNA Extraction, Genome Sequencing, Assembly and Annotation
2.4.2. Identification of Bacteriocin-Encoding Gene Cluster
2.4.3. Genotypic and Phenotypic Characterization of Probiotic Functions
2.5. Statistical Analysis
2.6. Genbank Accession Numbers
3. Results and Discussion
3.1. Diversity of LAB in Pancetta and Prosciutto
3.2. Organic Acid Production and Antagonistic Activity
3.3. Bacteriocin Activity and Associated Gene Clusters
3.4. Antibiotic Susceptibility and Screening for Probiotic Candidates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Binda, S.; Hill, C.; Johansen, E.; Obis, D.; Pot, B.; Sanders, M.E.; Tremblay, A.; Ouwehand, A.C. Criteria to qualify microorganisms as “probiotic” in foods and dietary supplements. Front. Microbiol. 2020, 11, 1662. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Girones, R.; Herman, L.; Koutsoumanis, K.; Lindqvist, R.; Nørrung, B.; et al. Scientific opinion on the update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA. EFSA J. 2017, 15, 3. [Google Scholar] [CrossRef] [Green Version]
- Pessoa, W.F.B.; Melgaço, A.C.C.; de Almeida, M.E.; Ramos, L.P.; Rezende, R.P.; Romano, C.C. In vitro activity of Lactobacilli with probiotic potential isolated from cocoa fermentation against Gardnerella vaginalis. BioMed Res. Int. 2017, 2017, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falah, F.; Vasiee, A.; Behbahani, B.A.; Yazdi, F.T.; Moradi, S.; Mortazavi, S.A.; Roshanak, S. Evaluation of adherence and anti-infective properties of probiotic Lactobacillus fermentum strain 4-17 against Escherichia coli causing urinary tract infection in humans. Microb. Pathog. 2019, 131, 246–253. [Google Scholar] [CrossRef]
- Choi, A.-R.; Patra, J.K.; Kim, W.J.; Kang, S.-S. Antagonistic activities and probiotic potential of lactic acid bacteria derived from a plant-based fermented food. Front. Microbiol. 2018, 9, 1963. [Google Scholar] [CrossRef]
- Bin Masalam, M.S.; Bahieldin, A.; Alharbi, M.G.; Al-Masaudi, S.; Al-Jaouni, S.K.; Harakeh, S.M.; Al-Hindi, R.R. Isolation, molecular characterization and probiotic potential of lactic acid bacteria in saudi raw and fermented milk. Evid. Based Complement. Altern. Med. 2018, 2018, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Munekata, P.E.S.; Pateiro, M.; Zhang, W.; Domínguez, R.; Xing, L.; Fierro, E.M.; Lorenzo, J.M. Autochthonous probiotics in meat products: Selection, identification, and their use as starter culture. Microorganisms 2020, 8, 1833. [Google Scholar] [CrossRef]
- Jampaphaeng, K.; Cocolin, L.; Maneerat, S. Selection and evaluation of functional characteristics of autochthonous lactic acid bacteria isolated from traditional fermented stinky bean (Sataw-Dong). Ann. Microbiol. 2016, 67, 25–36. [Google Scholar] [CrossRef]
- Rezac, S.; Kok, C.R.; Heermann, M.; Hutkins, R. Fermented foods as a dietary source of live organisms. Front. Microbiol. 2018, 9, 1785. [Google Scholar] [CrossRef]
- Urso, R.; Comi, G.; Cocolin, L. Ecology of lactic acid bacteria in Italian fermented sausages: Isolation, identification and molecular characterization. Syst. Appl. Microbiol. 2006, 29, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Van Ba, H.; Seo, H.-W.; Seong, P.-N.; Kang, S.-M.; Kim, Y.-S.; Cho, S.-H.; Park, B.-Y.; Ham, J.-S.; Kim, J.-H. Lactobacillus plantarum (KACC 92189) as a potential probiotic starter culture for quality improvement of fermented sausages. Food Sci. Anim. Resour. 2018, 38, 189–202. [Google Scholar] [CrossRef]
- Połka, J.; Rebecchi, A.; Pisacane, V.; Morelli, L.; Puglisi, E. Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 2015, 46, 342–356. [Google Scholar] [CrossRef]
- Geeraerts, W.; De Vuyst, L.; Leroy, F. Mapping the dominant microbial species diversity at expiration date of raw meat and processed meats from equine origin, an underexplored meat ecosystem, in the Belgian retail. Int. J. Food Microbiol. 2019, 289, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Federici, S.; Ciarrocchi, F.; Campana, R.; Ciandrini, E.; Blasi, G.; Baffone, W. Identification and functional traits of lactic acid bacteria isolated from Ciauscolo salami produced in central Italy. Meat Sci. 2014, 98, 575–584. [Google Scholar] [CrossRef]
- Kuley, E.; Özyurt, G.; Özogul, I.; Boga, M.; Akyol, I.; Rocha, J.M.; Özogul, F. The role of selected lactic acid bacteria on organic acid accumulation during wet and spray-dried fish-based silages. Contributions to the winning combination of microbial food safety and environmental sustainability. Microorganisms 2020, 8, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarman, A.A.; Ramli, N.N.; Ridla, M.; Yaman, M.A.; Setiyono, A.A. Effects of organic acids on salmonella enteritidis growth inhibition and ileum surface area in laying ducks fed anaerobically fermented feed. Int. J. Poult. Sci. 2017, 16, 98–104. [Google Scholar] [CrossRef]
- Kroöckel, L. The role of lactic acid bacteria in safety and flavour development of meat and meat products. In Lactic Acid Bacteria—R&D for Food, Health and Livestock Purposes; Kongo, M., Ed.; IntechOpen: London, UK, 2013. [Google Scholar]
- Corsetti, A.; Settanni, L.; Van Sinderen, D. Characterization of bacteriocin-like inhibitory substances (BLIS) from sourdough lactic acid bacteria and evaluation of their in vitro and in situ activity. J. Appl. Microbiol. 2004, 96, 521–534. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Vec, P.Å.; Vancanneyt, M.; Seman, M.; Snauwaert, C.; Lefebvre, K.; Sedláček, I.; Swings, J. Evaluation of (GTG)5-PCR for identification of Enterococcus spp. FEMS Microbiol. Lett. 2005, 247, 59–63. [Google Scholar] [CrossRef] [Green Version]
- Anisimova, E.A.; Yarullina, D.R. Antibiotic resistance of Lactobacillus strains. Curr. Microbiol. 2019, 76, 1407–1416. [Google Scholar] [CrossRef]
- Crowley, S.; Mahony, J.; Van Sinderen, D. Broad-spectrum antifungal-producing lactic acid bacteria and their application in fruit models. Folia Microbiol. 2012, 58, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Tagg, J.R.; McGiven, A.R. Assay system for bacteriocins. Appl. Microbiol. 1971, 21, 943. [Google Scholar] [CrossRef]
- Sathyabama, S.; Vijayabharathi, R.; Devi, P.B.; Kumar, M.R.; Priyadarisini, V.B. Screening for probiotic properties of strains isolated from feces of various human groups. J. Microbiol. 2012, 50, 603–612. [Google Scholar] [CrossRef]
- Shehata, M.; El Sohaimy, S.; El-Sahn, M.A.; Youssef, M. Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Ann. Agric. Sci. 2016, 61, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Mafra, J.F.; Cruz, A.I.C.; Santana, T.S.D.; Ferreira, M.A.; Araujo, F.M.; Evangelista-Barreto, N.S. Probiotic characterization of a commercial starter culture used in the fermentation of sausages. Food Sci. Technol. 2021, 41, 240–246. [Google Scholar] [CrossRef]
- Gilliland, S.; Staley, T.; Bush, L. Importance of bile tolerance of Lactobacillus acidophilus used as a dietary adjunct. J. Dairy Sci. 1984, 67, 3045–3051. [Google Scholar] [CrossRef]
- Melo, T.A.; Dos Santos, T.F.; Pereira, L.R.; Passos, H.M.; Rezende, R.P.; Romano, C.C. Functional profile evaluation of Lactobacillus fermentum TCUESC01: A new potential probiotic strain isolated during cocoa fermentation. BioMed Res. Int. 2017, 2017, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Vinderola, C.; Medici, M.; Perdigon, G. Relationship between interaction sites in the gut, hydrophobicity, mucosal immunomodulating capacities and cell wall protein profiles in indigenous and exogenous bacteria. J. Appl. Microbiol. 2004, 96, 230–243. [Google Scholar] [CrossRef]
- Lugli, G.A.; Milani, C.; Mancabelli, L.; Van Sinderen, D.; Ventura, M. MEGAnnotator: A user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevreux, B.; Wetter, T.; Suhai, S. Genome sequence assembly using trace signals and additional sequence information. In Proceedings of the German Conference on Bioinformatics (GCB), Hannover, Germany, 4–6 October 1999; pp. 45–56. [Google Scholar]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Punta, M.; Coggill, P.C.; Eberhardt, R.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2011, 40, D290–D301. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Dashkevicz, M.P.; Feighner, S.D. Development of a differential medium for bile salt hydrolase-active Lactobacillus spp. Appl. Environ. Microbiol. 1989, 55, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Even, S.; Lindley, N.D.; Loubière, P.; Cocaign-Bousquet, M. Dynamic response of catabolic pathways to autoacidification in Lactococcus lactis: Transcript profiling and stability in relation to metabolic and energetic constraints. Mol. Microbiol. 2002, 45, 1143–1152. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; de Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [Green Version]
- Guidone, A.; Zotta, T.; Ross, R.P.; Stanton, C.; Rea, M.; Parente, E.; Ricciardi, A. Functional properties of Lactobacillus plantarum strains: A multivariate screening study. LWT Food Sci. Technol. 2014, 56, 69–76. [Google Scholar] [CrossRef]
- Tran, K.T.; May, B.K.; Smooker, P.; Van, T.T.H.; Coloe, P.J. Distribution and genetic diversity of lactic acid bacteria from traditional fermented sausage. Food Res. Int. 2011, 44, 338–344. [Google Scholar] [CrossRef]
- Wanangkarn, A.; Liu, D.-C.; Swetwiwathana, A.; Jindaprasert, A.; Phraephaisarn, C.; Chumnqoen, W.; Tan, F.-J. Lactic acid bacterial population dynamics during fermentation and storage of Thai fermented sausage according to restriction fragment length polymorphism analysis. Int. J. Food Microbiol. 2014, 186, 61–67. [Google Scholar] [CrossRef]
- Albano, H.; Van Reenen, C.A.; Todorov, S.D.; Cruz, D.; Fraga, L.; Hogg, T.; Dicks, L.M.; Teixeira, P. Phenotypic and genetic heterogeneity of lactic acid bacteria isolated from “Alheira”, a traditional fermented sausage produced in Portugal. Meat Sci. 2009, 82, 389–398. [Google Scholar] [CrossRef]
- Mejri, L.; Hassouna, M. Characterization and selection of Lactobacillus plantarum species isolated from dry fermented sausage reformulated with camel meat and hump fat. Appl. Biol. Chem. 2016, 59, 533–542. [Google Scholar] [CrossRef]
- Parente, E.; Ciocia, F.; Ricciardi, A.; Zotta, T.; Felis, G.E.; Torriani, S. Diversity of stress tolerance in Lactobacillus plantarum, Lactobacillus pentosus and Lactobacillus paraplantarum: A multivariate screening study. Int. J. Food Microbiol. 2010, 144, 270–279. [Google Scholar] [CrossRef]
- Parlindungan, E.; May, B.K.; Jones, O.A.H. Metabolic insights into the effects of nutrient stress on Lactobacillus plantarum B21. Front. Mol. Biosci. 2019, 6, 75. [Google Scholar] [CrossRef] [Green Version]
- Heunis, T.; Deane, S.; Smit, S.; Dicks, L. Proteomic profiling of the acid stress response in Lactobacillus plantarum 423. J. Proteome Res. 2014, 13, 4028–4039. [Google Scholar] [CrossRef] [PubMed]
- Van Reckem, E.; Geeraerts, W.; Charmpi, C.; Van Der Veken, D.; De Vuyst, L.; Leroy, F. Exploring the link between the geographical origin of European fermented foods and the diversity of their bacterial communities: The case of fermented meats. Front. Microbiol. 2019, 10, 2302. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, S.; Papagianni, M.; Filiousis, G.; Ambrosiadis, I.; Koidis, P. Pediocin SA-1, an antimicrobial peptide from Pediococcus acidilactici NRRL B5627: Production conditions, purification and characterization. Bioresour. Technol. 2008, 99, 5384–5390. [Google Scholar] [CrossRef] [PubMed]
- Mandal, V.; Sen, S.K.; Mandal, N.C. Isolation and characterization of pediocin NV 5 producing Pediococcus acidilactici LAB 5 from vacuum-packed fermented meat product. Indian J. Microbiol. 2011, 51, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Morishita, Y.; Shiromizu, K. Characterization of Lactobacilli isolated from meats and meat products. Int. J. Food Microbiol. 1986, 3, 19–29. [Google Scholar] [CrossRef]
- Landeta, G.; Curiel, J.; Carrascosa, A.; Muñoz, R.; Rivas, B.D.L. Technological and safety properties of lactic acid bacteria isolated from Spanish dry-cured sausages. Meat Sci. 2013, 95, 272–280. [Google Scholar] [CrossRef] [Green Version]
- Domínguez-Manzano, J.; Jimenez-Diaz, R. Suppression of bacteriocin production in mixed-species cultures of lactic acid bacteria. Food Control 2013, 30, 474–479. [Google Scholar] [CrossRef]
- Peh, E.; Kittler, S.; Reich, F.; Kehrenberg, C. Antimicrobial activity of organic acids against Campylobacter spp. and development of combinations—A synergistic effect? PLoS ONE 2020, 15, e0239312. [Google Scholar] [CrossRef] [PubMed]
- Theron, M.M.; Lues, J.F. Organic acids and meat preservation: A review. Food Rev. Int. 2007, 23, 141–158. [Google Scholar] [CrossRef]
- Lepaus, B.M.; Rocha, J.S.; José, J.F.B.D.S. Organic acids and hydrogen peroxide can replace chlorinated compounds as sanitizers on strawberries, cucumbers and rocket leaves. Food Sci. Technol. 2020, 40, 242–249. [Google Scholar] [CrossRef] [Green Version]
- Arslan, S.; Eyi, A.; Özdemir, F. Spoilage potentials and antimicrobial resistance of Pseudomonas spp. isolated from cheeses. J. Dairy Sci. 2011, 94, 5851–5856. [Google Scholar] [CrossRef] [Green Version]
- Andreevskaya, M.; Jääskeläinen, E.; Johansson, P.; Ylinen, A.; Paulin, L.; Björkroth, J.; Auvinen, P. Food spoilage—Associated Leuconostoc, Lactococcus, and Lactobacillus species display different survival strategies in response to competition. Appl. Environ. Microbiol. 2018, 84, e00554-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oxaran, V.; Dittmann, K.K.; Lee, S.H.I.; Chaul, L.T.; Oliveira, C.; Corassin, C.H.; Alves, V.F.; De Martinis, E.C.P.; Gram, L. Behavior of foodborne pathogens Listeria monocytogenes and Staphylococcus aureus in mixed-species biofilms exposed to biocides. Appl. Environ. Microbiol. 2018, 84, e02038-18. [Google Scholar] [CrossRef] [Green Version]
- Miranda, R.C.; Schaffner, D.W. Longer contact times increase cross-contamination of Enterobacter aerogenes from surfaces to food. Appl. Environ. Microbiol. 2016, 82, 6490–6496. [Google Scholar] [CrossRef] [Green Version]
- Tannous, J.; Barda, O.; Luciano-Rosario, D.; Prusky, D.B.; Sionov, E.; Keller, N.P. New insight into pathogenicity and secondary metabolism of the plant pathogen Penicillium expansum through deletion of the epigenetic reader SntB. Front. Microbiol. 2020, 11, 610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavermicocca, P.; Valerio, F.; Visconti, A. Antifungal activity of phenyllactic acid against molds isolated from bakery products. Appl. Environ. Microbiol. 2003, 69, 634–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, P.O.D.S.D.; Mendonça, C.M.N.; Moreno, A.C.R.; Bueno, A.V.I.; De Almeida, S.R.Y.; Seibert, L.; Converti, A.; Watanabe, I.-S.; Gierus, M.; Oliveira, R.P.D.S. Antibacterial and antifungal activity of crude and freeze-dried bacteriocin-like inhibitory substance produced by Pediococcus pentosaceus. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Stoyanova, L.G.; Ustyugova, E.A.; Netrusov, A. Antibacterial metabolites of lactic acid bacteria: Their diversity and properties. Appl. Biochem. Microbiol. 2012, 48, 229–243. [Google Scholar] [CrossRef]
- Bédard, F.; Hammami, R.; Zirah, S.; Rebuffat, S.; Fliss, I.; Biron, E. Synthesis, antimicrobial activity and conformational analysis of the class IIa bacteriocin pediocin PA-1 and analogs thereof. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Golneshin, A.; Gor, M.-C.; Williamson, N.; Vezina, B.; Van, T.T.H.; May, B.K.; Smith, A.T. Discovery and characterisation of circular bacteriocin plantacyclin B21AG from Lactiplantibacillus plantarum B21. Heliyon 2020, 6, e04715. [Google Scholar] [CrossRef]
- Wang, Y.; Qin, Y.; Xie, Q.; Zhang, Y.; Hu, J.; Li, P. Purification and characterization of plantaricin LPL-1, a novel class IIa bacteriocin produced by Lactobacillus plantarum LPL-1 isolated from fermented fish. Front. Microbiol. 2018, 9, 2276. [Google Scholar] [CrossRef]
- Borrero, J.; Kelly, E.; O’Connor, P.M.; Kelleher, P.; Scully, C.; Cotter, P.; Mahony, J.; Van Sinderen, D. Plantaricyclin A, a novel circular bacteriocin produced by Lactobacillus plantarum NI326: Purification, characterization, and heterologous production. Appl. Environ. Microbiol. 2017, 84, e01801. [Google Scholar] [CrossRef] [Green Version]
- Gueimonde, M.; Sánchez, B.; Reyes-Gavilán, C.G.D.L.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef] [Green Version]
- Blandino, G.; Milazzo, I.; Fazio, D. Antibiotic susceptibility of bacterial isolates from probiotic products available in Italy. Microb. Ecol. Heal. Dis. 2008, 20, 199–203. [Google Scholar] [CrossRef] [Green Version]
- Kastner, S.; Perreten, V.; Bleuler, H.; Hugenschmidt, G.; Lacroix, C.; Meile, L. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst. Appl. Microbiol. 2006, 29, 145–155. [Google Scholar] [CrossRef]
- De Souza, B.M.S.; Borgonovi, T.F.; Casarotti, S.; Todorov, S.; Penna, A.L.B. Lactobacillus casei and Lactobacillus fermentum strains isolated from mozzarella cheese: Probiotic potential, safety, acidifying kinetic parameters and viability under gastrointestinal tract conditions. Probiotics Antimicrob. Proteins 2019, 11, 382–396. [Google Scholar] [CrossRef]
- Colloca, M.E.; Ahumada, M.C.; López, M.E.; Nader-Macías, M.E. Surface properties of Lactobacilli isolated from healthy subjects. Oral Dis. 2008, 6, 227–233. [Google Scholar] [CrossRef]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In vitro evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, H.C.; Melo, D.D.S.; Ramos, C.L.; Dias, D.R.; Schwan, R.F. Probiotic properties of Lactobacilli and their ability to inhibit the adhesion of enteropathogenic bacteria to Caco-2 and HT-29 cells. Probiotics Antimicrob. Proteins 2021, 13, 102–112. [Google Scholar] [CrossRef]
- Ng, S.Y.; Koon, S.S.; Padam, B.S.; Chye, F.Y. Evaluation of probiotic potential of lactic acid bacteria isolated from traditional Malaysian fermented Bambangan (Mangifera pajang). Cyta J. Food 2015, 13, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Succi, M.; Tremonte, P.; Reale, A.; Sorrentino, E.; Grazia, L.; Pacifico, S.; Coppola, R. Bile salt and acid tolerance of Lactobacillus rhamnosus strains isolated from Parmigiano Reggiano cheese. FEMS Microbiol. Lett. 2005, 244, 129–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasavid, P.; Chaiwatanarat, T.; Pusuwan, P.; Sritara, C.; Roysri, K.; Namwongprom, S.; Kuanrakcharoen, P.; Premprabha, T.; Chunlertrith, K.; Thongsawat, S.; et al. Normal solid gastric emptying values measured by scintigraphy using asian-style meal: A multicenter study in healthy volunteers. J. Neurogastroenterol. Motil. 2014, 20, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Degen, L.P.; Phillips, S.F. Variability of gastrointestinal transit in healthy women and men. Gut 1996, 39, 299–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdés, L.; Cuervo, A.; Salazar, N.; Ruas-Madiedo, P.; Gueimonde, M.; González, S. The relationship between phenolic compounds from diet and microbiota: Impact on human health. Food Funct. 2015, 6, 2424–2439. [Google Scholar] [CrossRef]
- Wood, J.M. Bacterial responses to osmotic challenges. J. Gen. Physiol. 2015, 145, 381–388. [Google Scholar] [CrossRef]
- Yang, X.; Hu, W.; Xiu, Z.; Jiang, A.; Sarengaowa, G.; Ji, Y.; Guan, Y.; Feng, K. Effect of salt concentration on microbial communities, physicochemical properties and metabolite profile during spontaneous fermentation of Chinese northeast sauerkraut. J. Appl. Microbiol. 2020, 129, 1458–1471. [Google Scholar] [CrossRef]
- Elias, M.; Laranjo, M.; Santos, A.C.A.; Potes, M.E. The role of salt on food and human health. Salt Earth 2020, 19. [Google Scholar] [CrossRef] [Green Version]
- Begley, M.; Hill, C.; Gahan, C.G.M. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Zhang, L.W.; Han, X.; Li, J.; Du, M.; Yi, H.; Feng, Z.; Zhang, Y.; Xu, X. Short communication: A sensitive method for qualitative screening of bile salt hydrolase-active lactobacilli based on thin-layer chromatography. J. Dairy Sci. 2011, 94, 1732–1737. [Google Scholar] [CrossRef]
- Devi, S.M.; Archer, A.C.; Halami, P.M. Screening, characterization and in vitro evaluation of probiotic properties among lactic acid bacteria through comparative analysis. Probiotics Antimicrob. Proteins 2015, 7, 181–192. [Google Scholar] [CrossRef]
- Meticulous Market Research. Probiotics Market by Strain (Bacillus), by Product Type (Dairy Food {Yogurt}, Baked Food}, Animal Feed), By Form (Liquid), By Sales Channel (Super Markets, Pharmacies), By End-User (Human {Adults, Senior}, Animal), and Geography- Global Forecast To 2027; Meticulous Market Research Pvt. Ltd.: Pune, India, 2021; p. 325. [Google Scholar]
- McFarland, L.V.; Evans, C.T.; Goldstein, E.J.C. Strain-specificity and disease-specificity of probiotic efficacy: A systematic review and meta-analysis. Front. Med. (Lausanne) 2018, 5, 124. [Google Scholar] [CrossRef]





| L. cremoris HP | L. paramesenteroides NCDO869 | L. mesenteroides NCDO2028 | K. aerogenes NCIMB10102 | E. faecium NCIMB11508 | P. aeruginosa PA01 | L. innocua UCC3 | S. aureus NCDO949 | P. expansum DSM1282 | |
|---|---|---|---|---|---|---|---|---|---|
| L. plantarum 41G | + + + | + + | + | + | + | + | + + | + | + + + |
| L. plantarum 41E | + + + | + + | + | + | + | + | + + + | + | + + + |
| L. plantarum 41O | + + + | + + | + | + | + | + | + + + + | + + + + | + + + |
| L. plantarum 41P | + + + | + + | + | + + | + | + | + + + | + + + | + + + |
| L. plantarum 13A | + + + | + + | + | + | + + + | + + | + + + | + + + | + + + |
| L. plantarum 37F | + + + | + + | + | + | + + + | + | + + | + + + | + + |
| L. plantarum 38I | + + + | + + | + | + + | + + | + + + | + + + + | + + + | + + + |
| L. plantarum 37I | + + + | + + | + | + + | + + | + + | + + + | + + + | + + |
| L. sakei 41D | + | + | + | + | + | + | + + | + | − |
| L. sakei 42C | + | + | + | + | + | + | + + | + | − |
| L. sakei 38F | + | + | + | + | + + | + | + + + | + + | − |
| L. sakei 39F | + | + | + | + | + | + | + + + | + + | − |
| L. curvatus 40A | + | − | − | − | − | − | − | − | − |
| L. curvatus 41A | + | − | − | − | − | − | − | − | − |
| L. curvatus 15A | − | − | − | − | − | − | − | + | − |
| L. curvatus 15E | − | − | − | − | − | − | − | + | − |
| L. coryniformis subsp. torquens 42L | + + | − | − | + | + | + | + + | + + | + |
| L. coryniformis subsp. torquens 42M | + | − | − | + | + + | + | + + | + + + | + |
| L. coryniformis subsp. torquens 14I | − | − | − | − | + | + | + | + | + |
| L. coryniformis subsp. torquens 15I | − | − | − | − | + | + + | + + | + + | + |
| P. acidilactici 40J | + | − | + + + + | + | + + + + | + | + + + + | + + + + | + + |
| P. acidilactici 40I | + | − | + + + + | + + | + + + + | + | + + + + | + + + | + + |
| Indicators | 13A | 41G | 38I | 41P | 40J |
|---|---|---|---|---|---|
| L. cremoris HP a | + | + | + | + | − |
| L. cremoris MG1363 d | + | + | + | + | − |
| L. cremoris NZ9000 d | + | + | + | + | − |
| L. paramesenteroides NCDO869 b | + | + | + | + | − |
| L. mesenteroides NCDO2028 b | − | − | − | − | + |
| K. aerogenes NCIMB10102 c | − | − | − | − | − |
| E. faecium NCIMB11508 c | − | − | − | − | + |
| P. aeruginosa PA01 a | − | − | − | − | − |
| L. innocua UCC3 d | − | − | − | − | + |
| L. innocua DPC3565 d | − | − | − | − | + |
| L. innocua DPC3566 d | − | − | − | − | + |
| L. innocua DPC3567 d | − | − | − | − | + |
| S. aureus NCDO949 b | − | − | − | − | − |
| Penicillin | Oxacilin | Ampicillin | Vancomycin | Gentamicin | Tetracycline | Erythromycin | Streptomycin | Chloramphenicol | Mupirocin | Rifampicin | Nalidixic Acid | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. curvatus 40A | S | R | S | R | R | S | S | R | R | S | S | R |
| L. curvatus 41A | R | R | S | R | S | S | S | R | S | S | S | R |
| L. curvatus 15E | S | R | S | R | R | S | S | R | S | S | S | R |
| L. curvatus 15A | S | R | S | R | R | S | S | R | S | S | S | R |
| L. sakei 41D | MS | R | S | R | R | S | S | R | R | S | S | R |
| L. sakei 42C | S | R | S | R | R | S | S | R | S | S | S | R |
| L. sakei 38F | R | R | S | R | R | S | S | R | R | S | S | R |
| L. sakei 39F | R | R | S | R | R | S | S | R | R | S | S | R |
| L. plantarum 37I | R | R | S | R | R | S | R | R | S | S | S | R |
| L. plantarum 38 I | R | R | S | R | R | S | S | R | S | S | R | R |
| L. plantarum 13A | R | R | S | R | R | S | S | R | R | S | R | R |
| L. plantarum 37F | R | R | S | R | R | S | S | R | MS | S | S | R |
| L. plantarum 41O | R | R | S | R | R | S | R | R | R | S | S | R |
| L. plantarum 41G | R | R | S | R | R | S | S | R | S | S | S | R |
| L. plantarum 41E | R | R | S | R | R | S | R | R | MS | S | R | R |
| L. plantarum 41P | R | R | S | R | R | S | S | R | R | S | S | R |
| P. acidilactici 40J | S | R | S | R | R | S | S | R | MS | S | S | R |
| P. acidilactici 40I | S | R | S | R | R | S | R | R | R | S | S | R |
| L. coryniformis subsp. torquens 42L | S | R | S | R | S | S | S | MS | S | S | S | R |
| L. coryniformis subsp. torquens 42M | S | R | S | R | S | S | R | MS | S | S | S | R |
| L. coryniformis subsp. torquens 14I | S | R | S | R | S | S | S | R | S | S | S | R |
| L. coryniformis subsp. torquens 15I | S | R | S | R | S | R | R | R | S | S | S | R |
| Strain | Hydrophobicity (%) | Autoaggregation (%) | ||
|---|---|---|---|---|
| 3 h | 5 h | 24 h | ||
| L. curvatus 41A | 8.2 ± 0.6 | 10.5 ± 0.4 | 13.9 ± 1.1 | 28.9 ± 1.5 |
| L. curvatus 15E | 7.8 ± 0.7 | 10.2 ± 0.9 | 12.3 ± 1.3 | 32.0 ± 6.2 |
| L. curvatus 15A | 18.1 ± 2.4 | 9.7 ± 2.9 | 15.1 ± 1.4 | 50.7 ± 5.8 |
| L. sakei 42C | 4.9 ± 0.5 | 12.6 ± 0.3 | 14.0 ± 0.4 | 45.1 ± 5.0 |
| L. plantarum 38I | 37.1 ± 0.3 | 11.4 ± 0.1 | 19.0 ± 1.0 | 66.9 ± 5.2 |
| L. plantarum 13A | 34.5 ± 0.4 | 10.6 ± 0.7 | 18.1 ± 0.4 | 56.9 ± 2.8 |
| L. plantarum 37F | 31.2 ± 0.5 | 11.8 ± 0.3 | 20.4 ± 0.9 | 63.6 ± 2.2 |
| L. plantarum 41G | 37.4 ± 0.7 | 11.9 ± 0.4 | 19.7 ± 0.7 | 67.6 ± 4.1 |
| L. plantarum 41P | 39.9 ± 2.0 | 11.4 ± 0.1 | 19.5 ± 0.4 | 69.0 ± 4.2 |
| P. acidilactici 40J | 11.5 ± 1.4 | 10.6 ± 0.7 | 14.4 ± 0.8 | 28.7 ± 3.8 |
| L. coryniformis subsp. torquens 42L | 38.8 ± 2.3 | 15.4 ± 1.1 | 22.5 ± 0.7 | 48.0 ± 3.6 |
| L.coryniformis subsp. torquens 14I | 42.2 ± 0.7 | 15.6 ± 1.3 | 22.5 ± 0.6 | 52.6 ± 4.5 |
| Strain | Mean ± SD (log10 CFU/mL) | |||
|---|---|---|---|---|
| 0 h | 1 h | 2 h | 3 h | |
| L. curvatus 41A | 8.16 ± 0.17 | − | − | − |
| L. curvatus 15E | 7.62 ± 0.45 | − | − | − |
| L. curvatus 15A | 7.53 ± 0.59 | − | − | − |
| L. sakei 42C | 7.46 ± 0.23 | − | − | − |
| L. plantarum 38I | 9.45 ± 0.02 | 8.03 ± 0.41 | 3.96 ± 0.74 | 2.09 ± 0.13 |
| L. plantarum 13A | 9.40 ± 0.20 | 7.94 ± 0.43 | 4.32 ± 0.82 | 3.11 ± 0.97 |
| L. plantarum 37F | 9.57 ± 0.11 | 7.90 ± 0.62 | 4.41 ± 0.42 | 2.98 ± 0.35 |
| L. plantarum 41G | 9.42 ± 0.22 | 7.84 ± 0.43 | 3.59 ± 1.21 | 2.00 ± 1.41 |
| L. plantarum 41P | 9.34 ± 0.20 | 7.10 ± 1.17 | 5.78 ± 0.55 | 3.31 ± 0.93 |
| P. acidilactici 40J | 9.56 ± 0.06 | 5.57 ± 0.42 | 4.40 ± 0.38 | 3.99 ± 0.43 |
| L. coryniformis subsp. torquens 42L | 8.79 ± 0.24 | 2.69 ± 0.53 | − | − |
| L.coryniformis subsp. torquens 14I | 8.40 ± 0.44 | 2.91 ± 0.20 | − | − |
| Strain | Mean ± SD (log10 CFU/mL) | |||
|---|---|---|---|---|
| 0 h | 1 h | 2 h | 3 h | |
| L. curvatus 41A | 7.59 ± 0.22 | 7.36 ± 0.11 | 7.08 ± 0.14 | 6.72 ± 0.18 |
| L. curvatus 15E | 7.57 ± 0.20 | 7.21 ± 0.23 | 7.07 ± 0.11 | 6.83 ± 0.34 |
| L. curvatus 15A | 7.22 ± 0.32 | 7.08 ± 0.23 | 6.90 ± 0.20 | 6.64 ± 0.21 |
| L. sakei 42C | 7.41 ± 0.11 | 7.06 ± 0.03 | 6.96 ± 0.23 | 6.52 ± 0.14 |
| L. plantarum 38I | 8.90 ± 0.18 | 8.77 ± 0.14 | 8.57 ± 0.20 | 8.41 ± 0.08 |
| L. plantarum 13A | 8.81 ± 0.03 | 8.86 ± 0.12 | 8.47 ± 0.14 | 8.22 ± 0.08 |
| L. plantarum 37F | 8.83 ± 0.15 | 8.70 ± 0.08 | 8.48 ± 0.16 | 8.23 ± 0.12 |
| L. plantarum 41G | 8.89 ± 0.15 | 8.81 ± 0.22 | 8.60 ± 0.10 | 8.38 ± 0.11 |
| L. plantarum 41P | 9.01 ± 0.08 | 8.61 ± 0.14* | 8.55 ± 0.14 | 8.39 ± 0.09 |
| P. acidilactici 40J | 9.01 ± 0.12 | 8.92 ± 0.04 | 8.48 ± 0.06 | 8.25 ± 0.05 |
| L. coryniformis subsp. torquens 42L | 8.59 ± 0.13 | 8.20 ± 0.26 | 7.94 ± 0.26 | 7.78 ± 0.26 |
| L.coryniformis subsp. torquens 14I | 8.73 ± 0.05 | 8.33 ± 0.40 | 8.09 ± 0.15 | 7.91 ± 0.22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parlindungan, E.; Lugli, G.A.; Ventura, M.; van Sinderen, D.; Mahony, J. Lactic Acid Bacteria Diversity and Characterization of Probiotic Candidates in Fermented Meats. Foods 2021, 10, 1519. https://doi.org/10.3390/foods10071519
Parlindungan E, Lugli GA, Ventura M, van Sinderen D, Mahony J. Lactic Acid Bacteria Diversity and Characterization of Probiotic Candidates in Fermented Meats. Foods. 2021; 10(7):1519. https://doi.org/10.3390/foods10071519
Chicago/Turabian StyleParlindungan, Elvina, Gabriele A. Lugli, Marco Ventura, Douwe van Sinderen, and Jennifer Mahony. 2021. "Lactic Acid Bacteria Diversity and Characterization of Probiotic Candidates in Fermented Meats" Foods 10, no. 7: 1519. https://doi.org/10.3390/foods10071519
APA StyleParlindungan, E., Lugli, G. A., Ventura, M., van Sinderen, D., & Mahony, J. (2021). Lactic Acid Bacteria Diversity and Characterization of Probiotic Candidates in Fermented Meats. Foods, 10(7), 1519. https://doi.org/10.3390/foods10071519