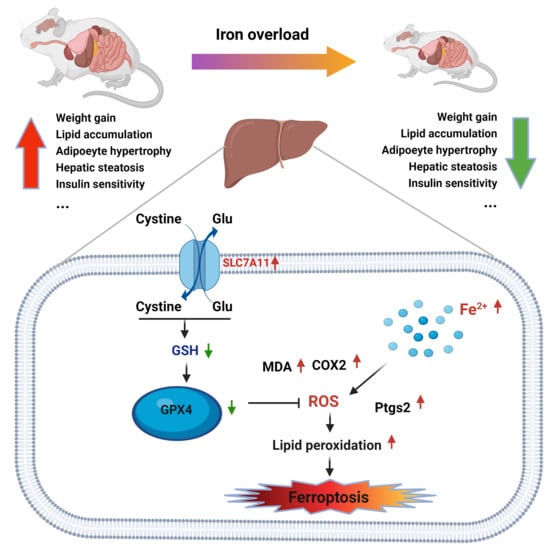
Iron Overload Protects from Obesity by Ferroptosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Experiments
2.2. Serum and Hepatic Biochemical Assays
2.3. Glucose and Insulin Tolerance Tests
2.4. Histological Analysis of the Liver
2.5. Real-Time Polymerase Chain Reaction (PCR) Analysis
2.6. Western Blot Analysis
2.7. RNA-Seq Analysis
2.8. Statistical Analyses
3. Results
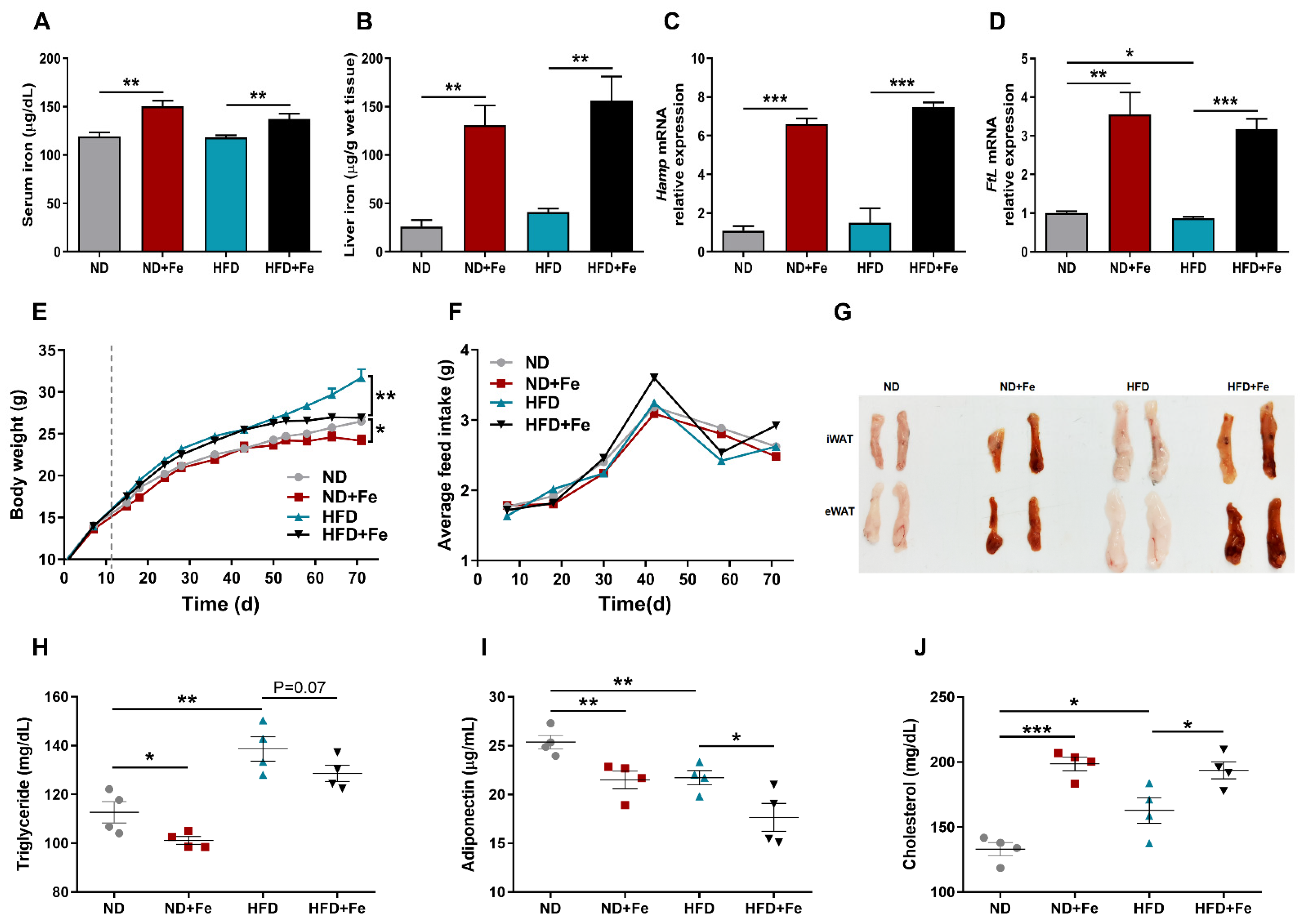
3.1. Iron Overload Protects against HFD-Induced Obesity
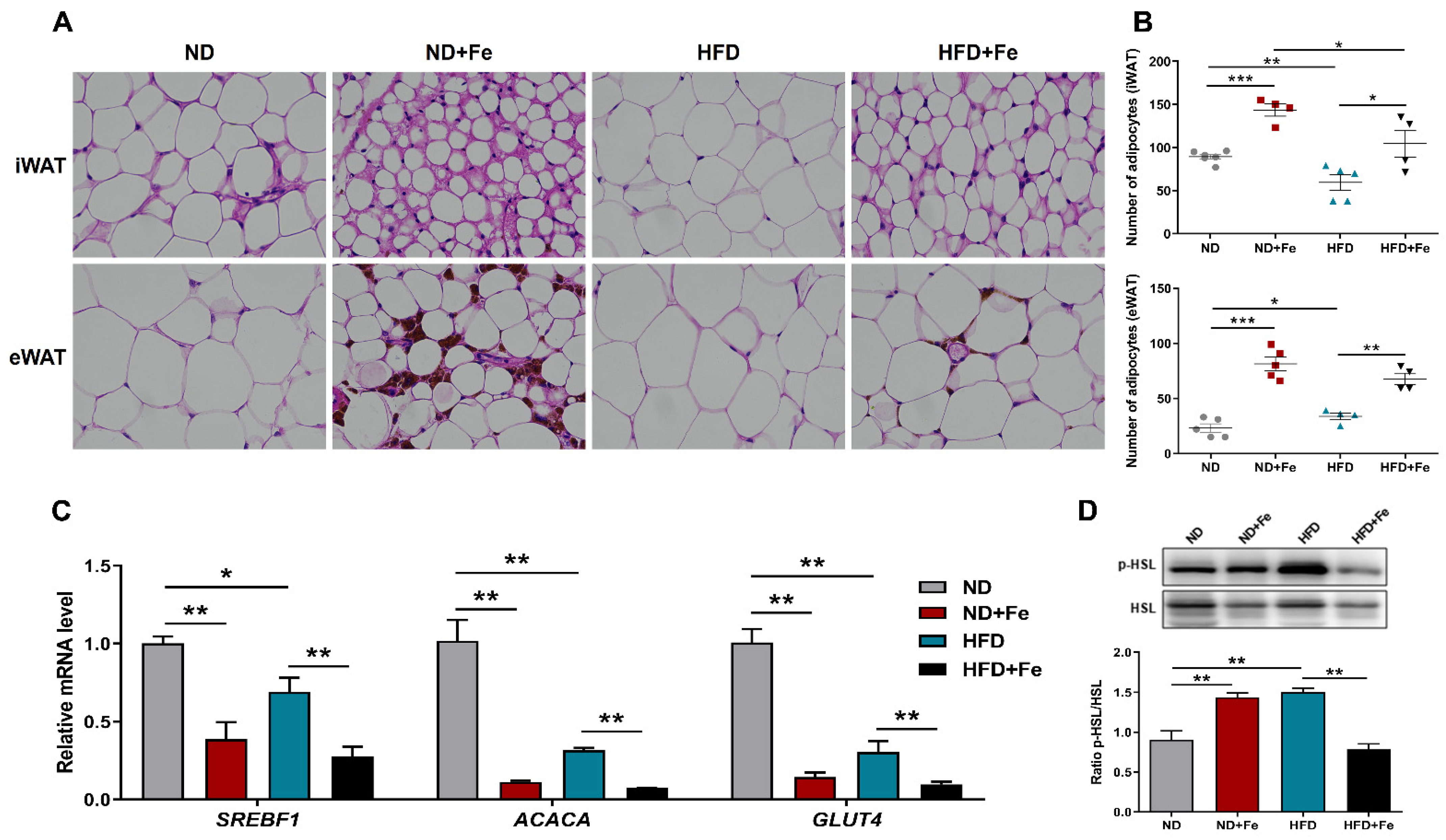
3.2. Reduced Adipocyte Hypertrophy Due to Iron Overload
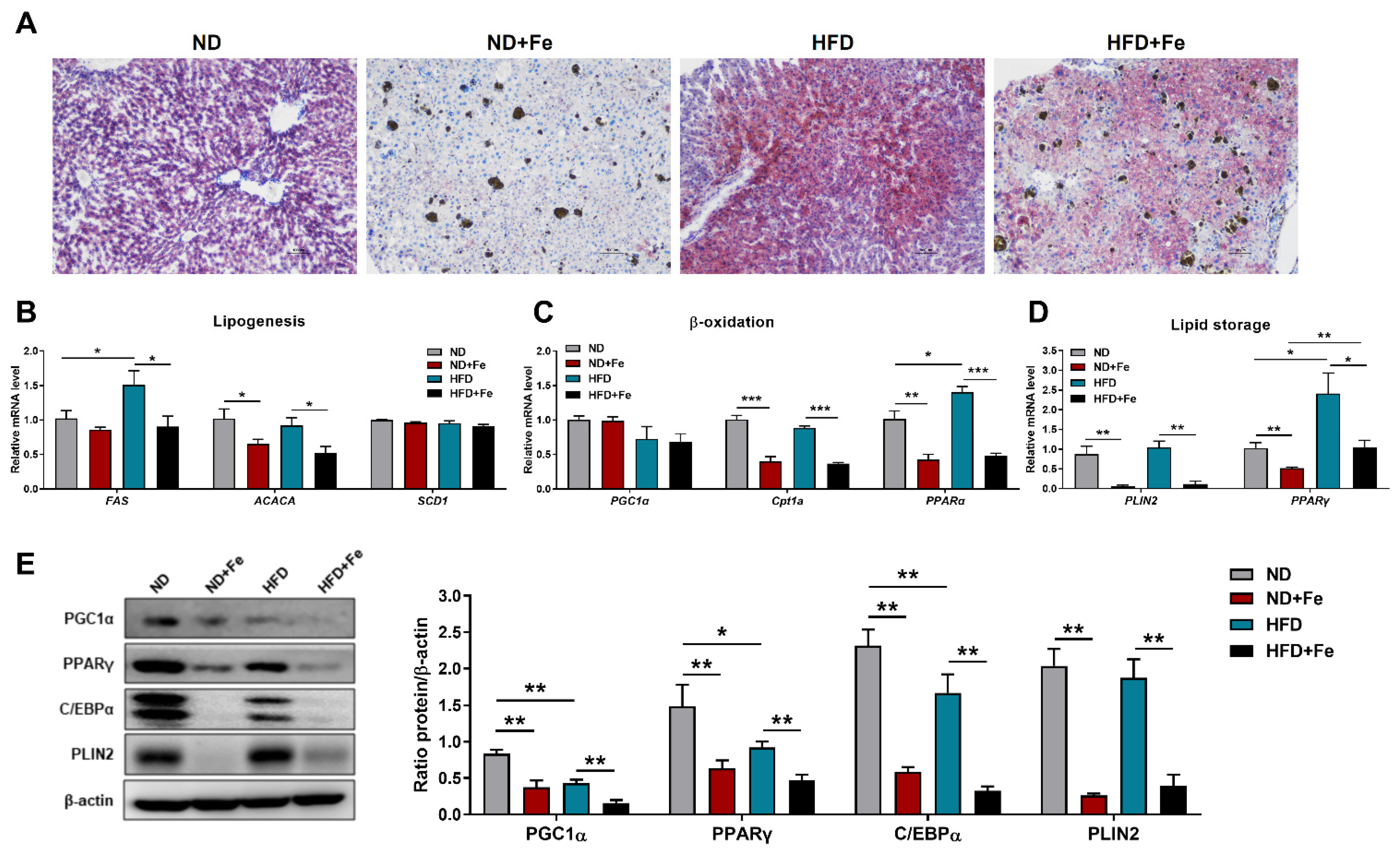
3.3. Decreased Lipidoses in the Liver of Iron-Overloaded Mice
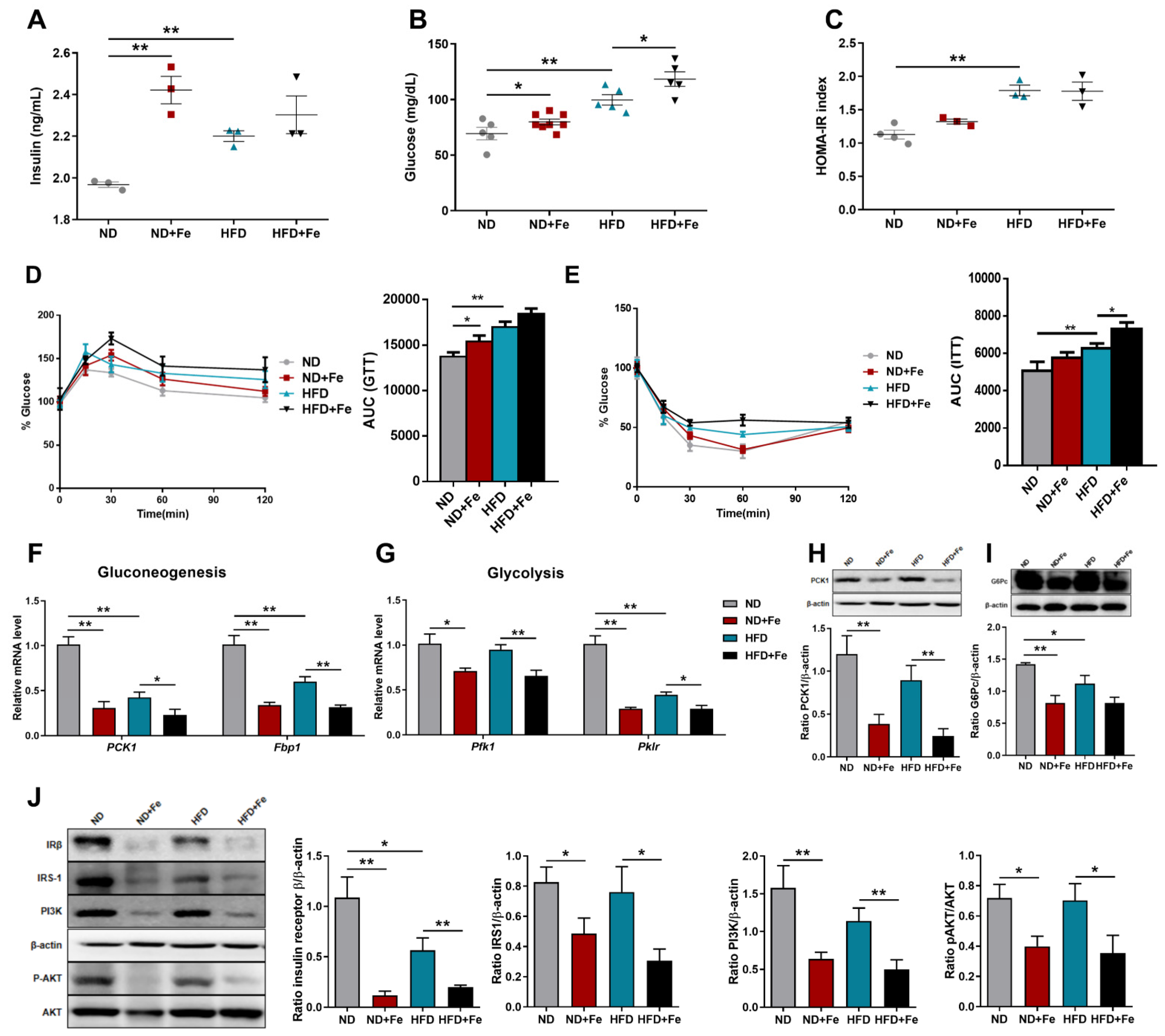
3.4. Decreased Insulin Sensitivity in Iron-Overloaded Mice
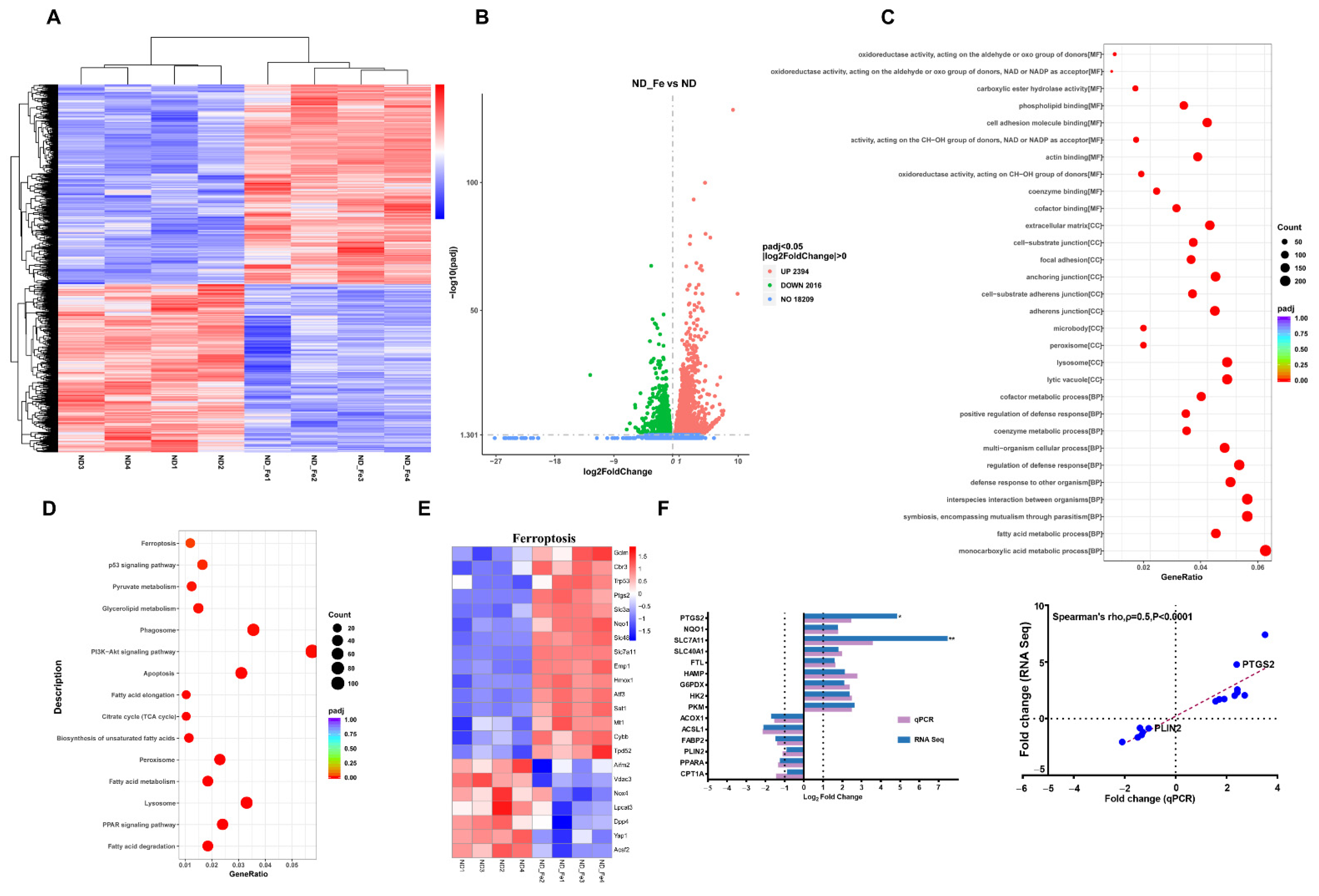
3.5. Enriched Ferroptosis and Fatty Acid Metabolism Pathways by Excess Iron
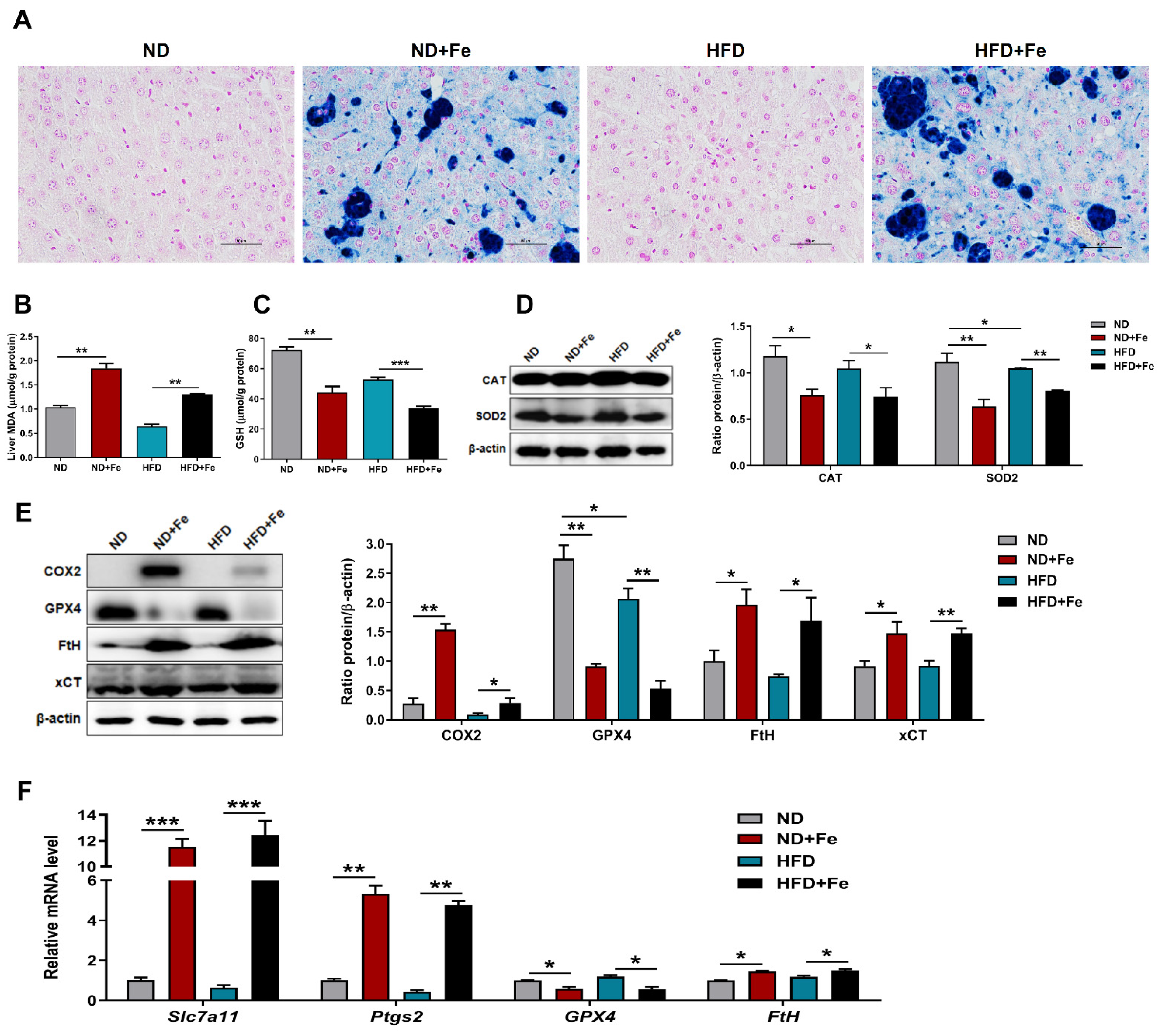
3.6. Induced Ferroptosis in the Liver of Iron Overload Mice
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Datz, C.; Muller, E.; Aigner, E. Iron overload and non-alcoholic fatty liver disease. Minerva. Endocrinol. 2017, 42, 173–183. [Google Scholar] [CrossRef]
- Wilson, J.G.; Lindquist, J.H.; Grambow, S.C.; Crook, E.D.; Maher, J.F. Potential role of increased iron stores in diabetes. Am. J. Med. Sci. 2003, 325, 332–339. [Google Scholar] [CrossRef]
- Pennell, D.J.; Udelson, J.E.; Arai, A.E.; Bozkurt, B.; Cohen, A.R.; Galanello, R.; Hoffman, T.M.; Kiernan, M.S.; Lerakis, S.; Piga, A.; et al. Cardiovascular function and treatment in beta-thalassemia major: A consensus statement from the American Heart Association. Circulation 2013, 128, 281–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brissot, P.; Loreal, O. Iron metabolism and related genetic diseases: A cleared land, keeping mysteries. J. Hepatol. 2016, 64, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Sikorska, K.; Bernat, A.; Wroblewska, A. Molecular pathogenesis and clinical consequences of iron overload in liver cirrhosis. Hepatobiliary Pancreat. Dis. Int. 2016, 15, 461–479. [Google Scholar] [CrossRef]
- Atarashi, M.; Izawa, T.; Miyagi, R.; Ohji, S.; Hashimoto, A.; Kuwamura, M.; Yamate, J. Dietary iron supplementation alters hepatic inflammation in a rat model of nonalcoholic steatohepatitis. Nutrients 2018, 10, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.; Jia, L.; Xiong, Q.; Feng, Y.; Du, H. The role of iron homeostasis in adipocyte metabolism. Food Funct. 2021, 12, 4246–4253. [Google Scholar] [CrossRef]
- Gabrielsen, J.S.; Gao, Y.; Simcox, J.A.; Huang, J.; Thorup, D.; Jones, D.; Cooksey, R.C.; Gabrielsen, D.; Adams, T.D.; Hunt, S.C.; et al. Adipocyte iron regulates adiponectin and insulin sensitivity. J. Clin. Investig. 2012, 122, 3529–3540. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Real, J.M.; Manco, M. Effects of iron overload on chronic metabolic diseases. Lancet Diabetes. Endocrinol. 2014, 2, 513–526. [Google Scholar] [CrossRef]
- Ma, W.; Feng, Y.; Jia, L.; Li, S.; Li, J.; Wang, Z.; Chen, X.; Du, H. Dietary iron modulates glucose and lipid homeostasis in diabetic mice. Biol. Trace Elem. Res. 2019, 189, 194–200. [Google Scholar] [CrossRef]
- Rasmussen, T.S.; Mentzel, C.; Kot, W.; Castro-Mejia, J.L.; Zuffa, S.; Swann, J.R.; Hansen, L.H.; Vogensen, F.K.; Hansen, A.K.; Nielsen, D.S. Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut 2020, 69, 2122–2130. [Google Scholar] [CrossRef]
- Vinue, A.; Gonzalez-Navarro, H. Glucose and Insulin Tolerance Tests in the Mouse. Methods Mol Biol. 2015, 1339, 247–254. [Google Scholar]
- Imamura, M.; Inoguchi, T.; Ikuyama, S.; Taniguchi, S.; Kobayashi, K.; Nakashima, N.; Nawata, H. ADRP stimulates lipid accumulation and lipid droplet formation in murine fibroblasts. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E775–E783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsusue, K.; Kusakabe, T.; Noguchi, T.; Takiguchi, S.; Suzuki, T.; Yamano, S.; Gonzalez, F.J. Hepatic steatosis in leptin-deficient mice is promoted by the PPARgamma target gene Fsp27. Cell Metab. 2008, 7, 302–311. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.D.; Hsu, C.H.; Wang, X.; Sakai, S.; Freeman, M.W.; Gonzalez, F.J.; Spiegelman, B.M. C/EBPalpha induces adipogenesis through PPARgamma: A unified pathway. Genes Dev. 2002, 16, 22–26. [Google Scholar] [CrossRef] [Green Version]
- Stechemesser, L.; Eder, S.K.; Wagner, A.; Patsch, W.; Feldman, A.; Strasser, M.; Auer, S.; Niederseer, D.; Huber-Schönauer, U.; Paulweber, B.; et al. Metabolomic profiling identifies potential pathways involved in the interaction of iron homeostasis with glucose metabolism. Mol. Metab. 2017, 6, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, H.; Palyha, O.; Mu, J. Restoration of insulin receptor improves diabetic phenotype in T2DM mice. JCI Insight 2019, 4, e124945. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; An, P.; Xie, E.; Wu, Q.; Fang, X.; Gao, H.; Zhang, Z.; Li, Y.; Wang, X.; Zhang, J.; et al. Characterization of ferroptosis in murine models of hemochromatosis. Hepatology 2017, 66, 449–465. [Google Scholar] [CrossRef]
- Valenzuela, R.; Rincon-Cervera, M.A.; Echeverria, F.; Barrera, C.; Espinosa, A.; Hernandez-Rodas, M.C.; Ortiz, M.; Valenzuela, A.; Videla, L.A. Iron-induced pro-oxidant and pro-lipogenic responses in relation to impaired synthesis and accretion of long-chain polyunsaturated fatty acids in rat hepatic and extrahepatic tissues. Nutrition 2018, 45, 49–58. [Google Scholar] [CrossRef]
- Folgueras, A.R.; Freitas-Rodriguez, S.; Ramsay, A.J.; Garabaya, C.; Rodriguez, F.; Velasco, G.; Lopez-Otin, C. Matriptase-2 deficiency protects from obesity by modulating iron homeostasis. Nat. Commun. 2018, 9, 1350. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Babitt, J.L. Liver iron sensing and body iron homeostasis. Blood 2019, 133, 18–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, L.W.; Dixon, J.L.; Hewett, D.G. Role of early case detection by screening relatives of patients with HFE-associated hereditary haemochromatosis. Best. Pract. Res. Clin. Haematol. 2005, 18, 221–234. [Google Scholar] [CrossRef]
- Abraham, D.; Rogers, J.; Gault, P.; Kushner, J.P.; McClain, D.A. Increased insulin secretory capacity but decreased insulin sensitivity after correction of iron overload by phlebotomy in hereditary haemochromatosis. Diabetologia 2006, 49, 2546–2551. [Google Scholar] [CrossRef] [Green Version]
- Creighton, M.T.; McClain, D.A. Diabetes and hemochromatosis. Curr. Diab. Rep. 2014, 14, 488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, Y.; Deng, X.; Xu, Y.; Gao, Z.; Li, H. Iron overload-induced rat liver injury: Involvement of protein tyrosine nitration and the effect of baicalin. Eur. J. Pharmacol. 2012, 680, 95–101. [Google Scholar] [CrossRef]
- Garcia-Perez, C.; Roy, S.S.; Naghdi, S.; Lin, X.; Davies, E.; Hajnoczky, G. Bid-induced mitochondrial membrane permeabilization waves propagated by local reactive oxygen species (ROS) signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 4497–4502. [Google Scholar] [CrossRef] [Green Version]
- Shimada, K.; Hayano, M.; Pagano, N.C.; Stockwell, B.R. Cell-Line Selectivity Improves the Predictive Power of Pharmacogenomic Analyses and Helps Identify NADPH as Biomarker for Ferroptosis Sensitivity. Cell. Chem. Biol. 2016, 23, 225–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedmann, A.J.; Schneider, M.; Proneth, B.; Tyurina, Y.Y.; Tyurin, V.A.; Hammond, V.J.; Herbach, N.; Aichler, M.; Walch, A.; Eggenhofer, E.; et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell. Biol. 2014, 16, 1180–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsurusaki, S.; Tsuchiya, Y.; Koumura, T.; Nakasone, M.; Sakamoto, T.; Matsuoka, M.; Imai, H.; Yuet-Yin, K.C.; Okochi, H.; Nakano, H.; et al. Hepatic ferroptosis plays an important role as the trigger for initiating inflammation in nonalcoholic steatohepatitis. Cell. Death. Dis. 2019, 10, 449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louandre, C.; Ezzoukhry, Z.; Godin, C.; Barbare, J.C.; Maziere, J.C.; Chauffert, B.; Galmiche, A. Iron-dependent cell death of hepatocellular carcinoma cells exposed to sorafenib. Int. J. Cancer 2013, 133, 1732–1742. [Google Scholar] [CrossRef] [PubMed]
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Hamp | TTGCGATACCAATGCAGAAG | TGCAACAGATACCACACTGG |
| FtL | CACCTACCTCTCTCTGGGCT | CGCGATCGTTCTGAAACTCG |
| SREBF1 | ACTTTTCCTTAACGTGGGCCT | CATCTCGGCCAGTGTCTGTT |
| ACACA | CTGTATGAGAAAGGCTATGTG | AACCTGTCTGAAGAGGTTAG |
| GLUT4 | TGAAGATGAGTGTCCTGTGCTG | GAAGTGCAAAGGGTGAGTGAG |
| FAS | TCCAAGACTGACTCGGCTACTGAC | GCAGCCAGGTTCGGAATGCTATC |
| SCD1 | GTGGGGTAATTATTTGTGACC | TTTTTCCCAGACAGTACAAC |
| PGC1α | TCCTCTTCAAGATCCTGTTA | CACATACAAGGGAGAATTGC |
| Cpt1α | GGGAGGAATACATCTACCTG | GAAGACGAATAGGTTTGAG |
| PPARα | AGTTCGGGAACAAGACGTTG | CAGTGGGGAGAGAGGACAGA |
| PLIN2 | GACACCACCTGCATGGCT | TGAAGCAGGGCCACTCTC |
| PPARγ | TCTTCCATCACGGAGAGGTC | GATGCACTGCCTATGAGCAC |
| PCK1 | AAGCGGATATGGTGGGAACTC | CTCCAAATTTCCTCCCAGGGT |
| Fbp1 | AAGTACTGATGAGCCTTCTG | GCTCACCATAATGAATTCTCC |
| Pfk1 | AAGAGACTGATTTTGAGCAC | CTCAGAAACCCTTGTCTATG |
| Pklr | GTGAAGAAGTTTGATGAGATCC | CAAGAAAACCTTCTCTGCTG |
| Slc7a11 | CTTTGTTGCCCTCTCCTGCTTC | CAGAGGAGTGTGCTTGTGGACA |
| Ptgs2 | GGGTTGCTGGGGGAAGAAA | CTCTGCTCTGGTCAATGGAGG |
| GPX4 | CCTCCCCAGTACTGCAACAG | GGCTGAGAATTCGTGCATGG |
| FtH | TCAGTCACTACTGGAACTGC | CGTGGTCACCCAGTTCTTTA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, W.; Jia, L.; Xiong, Q.; Du, H. Iron Overload Protects from Obesity by Ferroptosis. Foods 2021, 10, 1787. https://doi.org/10.3390/foods10081787
Ma W, Jia L, Xiong Q, Du H. Iron Overload Protects from Obesity by Ferroptosis. Foods. 2021; 10(8):1787. https://doi.org/10.3390/foods10081787
Chicago/Turabian StyleMa, Wan, Li Jia, Qingqing Xiong, and Huahua Du. 2021. "Iron Overload Protects from Obesity by Ferroptosis" Foods 10, no. 8: 1787. https://doi.org/10.3390/foods10081787
APA StyleMa, W., Jia, L., Xiong, Q., & Du, H. (2021). Iron Overload Protects from Obesity by Ferroptosis. Foods, 10(8), 1787. https://doi.org/10.3390/foods10081787