Detection of Enteric Viruses and Core Microbiome Analysis in Artisanal Colonial Salami-Type Dry-Fermented Sausages from Santa Catarina, Brazil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Colonial Salami-Type Dry-Fermented Sausage Production
2.2. Physical-Chemical Evaluation
2.3. Microbiological Analyses
2.4. Viral Detection by RT-qPCR
2.5. Metagenomic Analyses
2.6. Statistical Analyses
3. Results and Discussion
3.1. Physical-Chemical Characteristics
3.2. Viral Detection
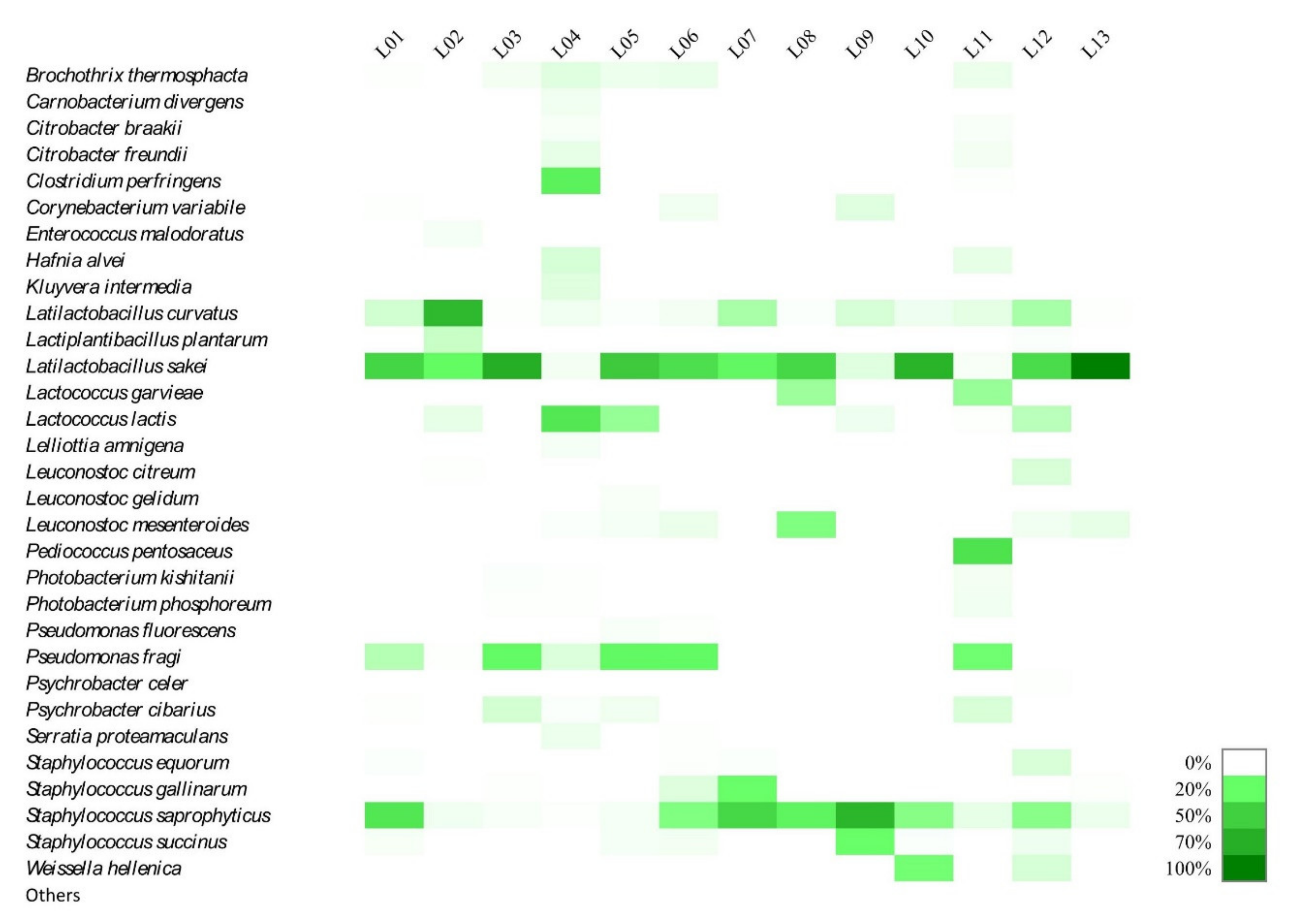
3.3. Microbiological and Metagenomic Analyses
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Toldrá, F.; Nip, W.K.; Hiu, Y.H. Dry-Fermented Sausages: An Overview. In Handbook of Fermented Meat and Poultry, 1st ed.; Toldrá, F., Ed.; Blackwell Publishing: Hoboken, NJ, USA, 2007; pp. 321–325. ISBN 0–8247–4780–1. [Google Scholar]
- Degenhardt, R.; Sant’ Anna, E.S. Pesquisa de Listeria sp, em embutidos cárneos fermentados produzidos na região Meio-Oeste de Santa Catarina, Brasil. Boletim do Centro de Pesquisa de Processamento de Alimentos 2007, 25, 133–140. [Google Scholar] [CrossRef]
- Fontana, C.; Fadda, S.; Cocconcelli, P.S.; Vignolo, G. Lactic Acid Bacteria in Meat Fermentations. In Lahtinen, 4th ed.; Ouwehand, A.C., Salminen, S., Von Wright, A., Eds.; CRC Press. Taylor & Francis Group: London, UK, 2012; pp. 247–264. [Google Scholar]
- Fraqueza, M.J. Antibiotic resistance of lactic acid bacteria isolated from dry-fermented sausages. Int. J. Food Microbiol. 2015, 212, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, L.; Zhang, H.; Wang, Y.; Chen, Q.; Kong, B. Physicochemical properties and flavour profile of fermented dry sausages with a reduction of sodium chloride. LWT 2020, 124, 109061. [Google Scholar] [CrossRef]
- Palavecino Prpich, N.Z.; Camprubí, G.E.; Cayré, M.E.; Castro, M.P. Indigenous Microbiota to Leverage Traditional Dry Sausage Production. Int. J. Food Sci. 2021, 30, 6696856. [Google Scholar] [CrossRef]
- Dalla Santa, O.R.; Chacón Alvarez, D.; Dalla Santa, H.S.; Zanette, C.M.; Freitas, R.J.S.; Macedo, R.E.F.; Terra, N.N. Microbiota of sausages obtained by spontaneous fermentation produced in the South of Brazil. Food Sci. Technol. 2012, 32, 653–660. [Google Scholar] [CrossRef] [Green Version]
- Aquilanti, L.; Garofalo, C.; Osimani, A.; Clementi, F. Ecology of lactic acid bacteria and coagulase negative cocci in fermented dry sausages manufactured in Italy and other Mediterranean countries: An overview. Int. Food Res. J. 2016, 23, 429–445. [Google Scholar]
- Cocolin, L.; Urso, R.; Rantsiou, K.; Cantoni, C.; Comi, G. Dynamics and characterization of yeasts during natural fermentation of Italian sausages. FEMS Yeast Res. 2006, 6, 692–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendonça, R.C.; Gouvêa, D.M.; Hungaro, H.M.; Sodré, A.D.F.; Querol-Simon, A. Dynamics of the yeast flora in artisanal country style and industrial dry cured sausage (yeast in fermented sausage). Food Control. 2013, 29, 143–148. [Google Scholar] [CrossRef] [Green Version]
- Sunesen, L.O.; Stahnke, L.H. Mould starter cultures for dry sausages—Selection, application and effects. Meat Sci. 2003, 65, 935–948. [Google Scholar] [CrossRef]
- Knipe, D.M.; Howley, P.M. (Eds.) Fields Virology, 6th ed.; Lippincott Williams & Wilkins a Wolters Kluwer Business: Philadelphia, PA, USA, 2013. [Google Scholar]
- Doceul, V.; Bagdassarian, E.; Demange, A.; Pavio, N. Zoonotic hepatitis E virus: Classification, animal reservoirs and transmission routes. Viruses 2016, 8, 270. [Google Scholar] [CrossRef]
- García, N.; Hernández, M.; Gutierrez-Boada, M.; Valero, A.; Navarro, A.; Muñoz-Chimeno, M.; Fernández-Manzano, A.; Escobar, F.M.; Martínez, I.; Bárcena, C.; et al. Occurrence of Hepatitis E Virus in Pigs and Pork Cuts and Organs at the Time of Slaughter, Spain. Front. Microbiol. 2020, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Doro, R.; Farkas, S.L.; Martella, V.; Banyai, K. Zoonotic transmission of rotavirus: Surveillance and control. Expert Rev. Anti-Infect. Ther. 2015, 13, 1337–1350. [Google Scholar] [CrossRef]
- Krog, J.S.; Hjulsager, C.K.; Larsen, M.A.; Larsen, L.E. Triple-reassortant influenza A virus with H3 of human seasonal origin, NA of swine origin, and internal A (H1N1) pandemic 2009 genes is established in Danish pigs. Influenza Other Respir. Viruses 2017, 11, 298–303. [Google Scholar] [CrossRef] [Green Version]
- Souza, D.S.M.; Tápparo, D.C.; Rogovski, P.; Cadamuro, R.D.; de Souza, E.B.; da Silva, R.; Degenhardt, R.; De Dea Lindner, J.; Viancelli, A.; Michelon, W.; et al. Hepatitis E Virus in Manure and Its Removal by Psychrophilic anaerobic Biodigestion in Intensive Production Farms, Santa Catarina, Brazil, 2018. Microorganisms 2020, 8, 2045. [Google Scholar] [CrossRef]
- Amorim, A.R.; Mendes, G.S.; Pena, G.P.A.; Santos, N. Hepatitis E virus infection of slaughtered healthy pigs in Brazil. Zoonoses Public Health 2018, 65, 501–504. [Google Scholar] [CrossRef]
- Tengan, F.M.; Figueiredo, G.M.; Nunes, A.K.; Manchiero, C.; Dantas, B.P.; Magri, M.C.; Prata, T.V.G.; Nascimento, M.; Mazza, C.C.; Abdala, E.; et al. Seroprevalence of hepatitis E in adults in Brazil: A systematic review and meta-analysis. Infect. Dis. Poverty 2019, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- BRASIL Ministério da Agricultura e do Abastecimento. Instrução Normativa n° 20, de 31 de julho de 2000, que oficializa os Métodos Analíticos Físico-químicos, para controle dos produtos cárneos e seus ingredientes–Sal e Salmoura. Diário Oficial da União, 27 jul. Seção 1, p. 10; Ministério da Agricultura e do Abastecimento: Brasília, Brazil, 2020. [Google Scholar]
- AOAC. Official Methods of Analysis; Association of Official Analytical Chemists: Gaithersburg, MD, USA, 2002. [Google Scholar]
- Koral, S.; Köse, S. The Effect of Using Frozen Raw Material and Different Salt Ratios on the Quality Changes of Dry Salted Atlantic Bonito (Lakerda) at Two Storage Conditions. Food Health 2018, 4, 213–230. [Google Scholar] [CrossRef]
- International Standards Organization. Microbiology of Food and Animal Feeding Stuffs–Horizontal Method for the Enumeration of Coliforms—Colony-Count Technique; ISO 4832:2012; International Standards Organization: Geneva, Switzerland, 2012. [Google Scholar]
- International Standards Organization. Microbiology of Food and Animal Feeding Stuffs–Horizontal Method for the Enumeration of Coagulase-Positive Staphylococci (Staphylococcus Aureus and Other Species) Part 1: Technique Using Baird-Parker Agar Medium; ISO 6888–1:2019; International Standards Organization: Geneva, Switzerland, 2019. [Google Scholar]
- International Standards Organization. Microbiology of Food and Animal Feeding Stuffs–Horizontal Method for the Enumeration of Sulfite-Reducing Bacteria Growing under Anaerobic Conditions; ISO 15213:2003; International Standards Organization: Geneva, Switzerland, 2003. [Google Scholar]
- International Standards Organization. Microbiology of Food and Animal Feeding Stuffs–Horizontal Method for the Enumeration of Mesophilic Lactic Acid Bacteria—Colony-Count Technique at 30 Degrees °C; ISO 15214:1998; International Standards Organization: Geneva, Switzerland, 1998. [Google Scholar]
- International Standards Organization. Microbiology of Food and Animal Feeding Stuffs–Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp.; ISO 6579–1:2017; International Standards Organization: Geneva, Switzerland, 2017. [Google Scholar]
- International Standards Organization. Microbiology of Food and Animal Feeding Stuffs–Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and of Listeria spp. Part 1: Detection Method; ISO 11290–1:2017; International Standards Organization: Geneva, Switzerland, 2017. [Google Scholar]
- International Standards Organization. Microbiology of Food and Animal Feeding Stuffs–Horizontal Method for the Enumeration of Yeasts and Moulds—Part 2: Colony Count Technique in Products with Water Activity Less Than or Equal to 0.95, ISO 21527–2:2008 standard.; International Standards Organization: Geneva, Switzerland, 2008. [Google Scholar]
- Rodríguez-Lázaro, D.; Cook, N.; Ruggeri, F.M.; Sellwood, J.; Nasser, A.; Nascimento, M.S.J.; van der Poel, W.H.M. Virus hazards from food, water and other contaminated environments. FEMS Microb. Rev. 2012, 36, 786–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baert, L.; Wobus, C.E.; Van Coillie, E.; Thackray, L.B.; Debevere, J.; Uyttendaele, M. Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Appl. Environ. Microbiol. 2008, 74, 543–546. [Google Scholar] [CrossRef] [Green Version]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.Q.; Halkosalo, A.; Salminen, M.; Szakal, E.D.; Puustinen, L.; Vesikari, T. One-step quantitative RT-PCR for the detection of rotavirus in acute gastroenteritis. J. Virol. Methods 2008, 153, 238–240. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Knight, R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Qian, P.Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 2009, 4, e7401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christoff, A.P.; Cruz, G.N.F.; Sereia, A.F.R.; Yamanaka, L.E.; Silveira, P.P.; De Oliveira, L.F.V. End-to-end assessment of fecal bacteriome analysis: From sample processing to DNA sequencing and bioinformatics results. BioRxiv 2019, 646349. [Google Scholar] [CrossRef]
- Schmidt, P.A.; Bálint, M.; Greshake, B.; Bandow, C.; Römbke, J.; Schmitt, I. Illumina metabarcoding of a soil fungal community. Soil Biol. Biochem. 2013, 65, 128–132. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Knight, R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- BRASIL Ministério da Agricultura e do Abastecimento. Instrução Normativa n°22, de 31 de julho de 2000, aprova os Regulamento Técnicos de Identidade e Qualidade de Copa, de Jerked Beef, de Presunto tipo Parma, de Presunto Cru, de Salame, de Salaminho, de Salaminho tipo Alemão, de Salame tipo Calabrês, de Salame tipo Friolano, de Salame tipo Napolitano, de Salame tipo Hamburguês, de Salame tipo Italiano, de Salame tipo Milano, de Lingüiça Colonial e Pepperoni. Diário Oficial da União, 3 ago. Seção 1, p. 15; Ministério da Agricultura e do Abastecimento: Brasília, Brazil, 2020.
- Ingham, S.C.; Engel, R.A.; Fanslau, M.A.; Schoeller, E.L.; Searls, G.; Buege, D.R.; Zhu, J. Fate of Staphylococcus aureus on Vacuum-Packaged Ready-to-Eat Meat Products Stored at 21 °C. J. Food Protec. 2005, 68, 1911–1915. [Google Scholar] [CrossRef] [Green Version]
- Tim, H. Sodium–Technological functions of salt in the manufacturing of food and drink products. Br. Food J. 2002, 104, 126–152. [Google Scholar] [CrossRef]
- De Souza, A.J.S.; Gomes-Gouvêa, M.S.; Soares, M.d.C.P.; Pinho, J.R.R.; Malheiros, A.P.; Carneiro, L.A.; Dos Santos, D.R.L.; Pereira, W.L.A. HEV infection in swine from Eastern Brazilian Amazon: Evidence of co-infection by different subtypes. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 477–485. [Google Scholar] [CrossRef]
- Boxman, I.L.; Jansen, C.C.; Zwartkruis-Nahuis, A.J.; Hägele, G.; Sosef, N.P.; Dirks, R.A. Detection and quantification of hepatitis E virus RNA in ready to eat raw pork sausages in the Netherlands. Int. J. Food Microb. 2020, 333, 108791. [Google Scholar] [CrossRef] [PubMed]
- Berto, A.; Grierson, S.; Hakze-van der Honing, R.; Martelli, F.; Johne, R.; Reetz, J.; Banks, M. HEV in Pork Liver sausages, France. Emerg. Infec. Dis. 2013, 19, 3–5. [Google Scholar] [CrossRef]
- Colson, P.; Borentain, P.; Queyriaux, B.; Kaba, M.; Moal, V.; Gallian, P.; Gerolami, R. Pig Liver Sausage as a Source of Hepatitis E Virus Transmission to Humans. J. Infect. Dis. 2010, 202, 825–834. [Google Scholar] [CrossRef] [Green Version]
- Wolff, A.; Günther, T.; Albert, T.; Schilling-Loeffler, K.; Gadicherla, A.K.; Johne, R. Stability of hepatitis E virus at different pH values. Int. J. Food Microbiol. 2020, 325, 108625. [Google Scholar] [CrossRef]
- Wolff, A.; Günther, T.; Albert, T.; Johne, R. Effect of Sodium Chloride, Sodium Nitrite and Sodium Nitrate on the Infectivity of Hepatitis E Virus. Food Environ. Virol. 2020, 12, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Chauhan, R.S.; Mahendran, M.; Malik, S.V.S. Rotavirus diarrhea in bovines and other domestic animals. Vet. Res. Commun. 2009, 33, 1–23. [Google Scholar] [CrossRef]
- Terzić-Vidojević, A.; Veljović, K.; Tolinački, M.; Živković, M.; Lukić, J.; Lozo, J.; Golić, N. Diversity of non-starter lactic acid bacteria in autochthonous dairy products from Western Balkan Countries-technological and probiotic properties. Food Res. Int. 2020, 136, 109494. [Google Scholar] [CrossRef]
- Lund, P.A.; De Biase, D.; Liran, O.; Scheler, O.; Mira, N.P.; Cetecioglu, Z.; O’Byrne, C. Understanding how microorganisms respond to acid pH is central to their control and successful exploitation. Front. Microbiol. 2020, 11, 2233. [Google Scholar] [CrossRef]
- Franciosa, I.; Alessandria, V.; Dolci, P.; Rantsiou, K.; Cocolin, L. Sausage fermentation and starter cultures in the era of molecular biology methods. Int. J. Food Microbiol. 2018, 279, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Hebert, E.M.; Saavedra, L.; Taranto, M.P.; Mozzi, F.; Christian Magni, C.; Nader, M.E.F.; de Valdez, G.F.; Sesma, F.; Vignolo, G.; Raya, R.R. Genome Sequence of the Bacteriocin-Producing Lactobacillus curvatus Strain CRL. J. Bacteriol. 2012, 194, 538–539. [Google Scholar] [CrossRef] [Green Version]
- Mainar, M.S.; Xhaferi, R.; Samapundo, S.; Devlieghere, F.; Leroy, F. Opportunities and limitations for the production of safe fermented meats without nitrate and nitrite using an antibacterial Staphylococcus sciuri starter culture. Food Control. 2016, 69, 267–274. [Google Scholar] [CrossRef]
- Colak, H.; Hampikyan, H.; Ulusoy, B.; Bingol, E.B. Presence of Listeria monocytogenes in Turkish style fermented sausage (sucuk). Food Control. 2007, 18, 30–32. [Google Scholar] [CrossRef]
- De Cesare, A.; Mioni, R.; Manfreda, G. Prevalence of Listeria monocytogenes in fresh and fermented Italian sausages and ribotyping of contaminating strains. Int. J. Food Microbiol. 2007, 120, 124–130. [Google Scholar] [CrossRef]
- Meloni, D. Presence of Listeria monocytogenes in Mediterranean-style dry fermented sausages. Foods 2015, 4, 34–50. [Google Scholar] [CrossRef] [Green Version]
- Auvolat, A.; Besse, N.G. The challenge of enumerating Listeria monocytogenes in food. Food Microb. 2016, 53, 135–149. [Google Scholar] [CrossRef]
- Thevenot, D.; Delignette-Muller, M.L.; Christieans, S.; Vernozy-Rozand, C. Prevalence of Listeria monocytogenes in 13 dried sausage processing plants and their products. Int. J. Food Microb. 2005, 102, 85–94. [Google Scholar] [CrossRef]
- Mainar, M.S.; Stavropoulou, D.A.; Leroy, F. Exploring the metabolic heterogeneity of coagulase-negative staphylococci to improve the quality and safety of fermented meats: A review. Int. J. Food Microb. 2017, 247, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Stellato, G.; La Storia, A.; De Filippis, F.; Borriello, G.; Villani, F.; Ercolini, D. Overlap of spoilage-associated microbiota between meat and the meat processing environment in small-scale and large-scale retail distributions. Appl. Environ. Microb. 2016, 82, 4045–4054. [Google Scholar] [CrossRef] [Green Version]
- Andrade, M.J.; Córdoba, J.J.; Casado, E.M.; Córdoba, M.G.; Rodríguez, M. Effect of selected strains of Debaryomyces hansenii on the volatile compound production of dry fermented sausage “salchichón”. Meat Sci. 2010, 85, 256–264. [Google Scholar] [CrossRef]
- Cano-García, L.; Rivera-Jiménez, S.; Belloch, C.; Flores, M. Generation of aroma compounds in a fermented sausage meat model system by Debaryomyces hansenii strains. Food Chem. 2014, 151, 364–373. [Google Scholar] [CrossRef]
- Núñez, F.; Lara, M.S.; Peromingo, B.; Delgado, J.; Sánchez-Montero, L.; Andrade, M.J. Selection and evaluation of Debaryomyces hansenii isolates as potential bioprotective agents against toxigenic penicillia in dry-fermented sausages. Food Microb. 2015, 46, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Encinas, J.P.; López-Díaz, T.M.; García-López, M.L.; Otero, A.; Moreno, B. Yeast populations on Spanish fermented sausages. Meat Sci. 2000, 54, 203–208. [Google Scholar] [CrossRef]
- Mižáková, A.; Pipová, M.; Turek, P. The occurrence of moulds in fermented raw meat products. Czech. J. Food Sci. 2002, 20, 89–94. [Google Scholar] [CrossRef] [Green Version]
- Ciegler, A.; Mintzlaff, H.J.; Weisleder, D.; Leistner, L. Potential production and detoxification of penicillic acid in mold-fermented sausage (salami). Appl. Microb. 1972, 24, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Castellari, C.; Quadrelli, A.M.; Laich, F. Surface mycobiota on Argentinean dry fermented sausages. Int. J. Food Microbiol. 2010, 142, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Sonjak, S.; Ličen, M.; Frisvad, J.C.; Gunde-Cimerman, N. The mycobiota of three dry-cured meat products from Slovenia. Food Microbiol. 2011, 28, 373–376. [Google Scholar] [CrossRef]
- Zadravec, M.; Vahčić, N.; Brnić, D.; Markov, K.; Frece, J.; Beck, R.; Pleadin, J. A study of surface moulds and mycotoxins in Croatian traditional dry-cured meat products. Int. J. Food Microbiol. 2020, 317, 108459. [Google Scholar] [CrossRef]

| Sample | City | Ripening (Days) | Casing | pH | Moisture (%) | NaCl (%) | WPS (%) | aw |
|---|---|---|---|---|---|---|---|---|
| L01 | Piratuba | 40 | N | 5.50 | 30.02 | 4.30 | 12.50 | 0.891 |
| L02 | Herval Velho | 16 | S | 6.30 | 50.53 | 3.70 | 6.80 | 0.950 |
| L03 | Jaborá | 1 | S | 6.10 | 51.14 | 3.17 | 5.80 | 0.957 |
| L04 | Concórdia | 7 | S | 5.10 | 30.20 | 6.15 | 16.90 | 0.829 |
| L05 | Luzerna | 2 | N | 5.80 | 50.22 | 4.20 | 7.70 | 0.943 |
| L06 | Videira | 9 | N | 5.50 | 41.03 | 4.32 | 9.50 | 0.927 |
| L07 | Salto Veloso | 12 | N | 6.40 | 40.85 | 4.75 | 10.40 | 0.917 |
| L08 | Salto Veloso | 6 | N | 5.60 | 38.10 | 3.01 | 7.30 | 0.946 |
| L09 | Tangará | 6 | N | 6.00 | 56.69 | 4.18 | 6.90 | 0.949 |
| L10 | Caçador | 16 | N | 6.00 | 28.48 | 5.04 | 15.00 | 0.857 |
| L11 | Videira | 2 | N | 6.00 | 43.22 | 4.49 | 9.40 | 0.928 |
| L12 | Iomerê | 10 | N | 5.70 | 39.02 | 4.92 | 11.20 | 0.908 |
| L13 | Lacerdópolis | 7 | S | 5.40 | 39.06 | 4.41 | 10.10 | 0.920 |
| Sample | TC * | TTC * | CPS * | SRC * | LAB * | Enterococci * | Yeasts * | Molds * | Salmonella ** | Listeria spp. ** |
|---|---|---|---|---|---|---|---|---|---|---|
| L01 | 2.04 | 2.00 | 2.00 | 1.00 | 7.60 | 5.89 | 8.34 | <2.00 | A | P |
| L02 | <1.00 | <1.00 | <2.00 | <1.00 | 6.91 | 4.29 | 6.64 | 7.00 | A | P |
| L03 | 1.70 | 1.70 | <2.00 | <1.00 | 8.32 | 3.11 | 3.11 | 7.30 | A | A |
| L04 | 3.89 | 3.81 | <2.00 | 1.48 | 6.60 | 5.68 | 6.81 | 4.30 | A | A |
| L05 | 4.73 | 4.73 | 2.48 | <1.00 | 7.84 | 5.78 | 8.30 | 7.48 | A | P |
| L06 | 2.26 | <1.00 | <2.00 | <1.00 | 7.84 | 3.63 | 7.28 | 7.00 | A | A |
| L07 | 1.00 | 1.00 | 4.73 | <1.00 | 7.86 | 6.20 | 6.81 | 7.30 | A | A |
| L08 | <1.00 | <1.00 | <2.00 | <1.00 | 7.91 | 3.67 | 6.78 | 7.00 | A | A |
| L09 | 2.08 | 1.60 | 2.90 | <1.00 | 8.38 | 6.25 | 7.20 | 4.48 | A | A |
| L10 | <1.00 | <1.00 | <2.00 | <1.00 | 6.79 | 3.15 | 6.55 | 7.30 | A | A |
| L11 | 2.86 | 2.28 | 2.00 | <1.00 | 6.75 | 4.70 | 7.47 | 7.30 | A | A |
| L12 | 2.00 | 2.00 | 2.30 | <1.00 | 8.48 | 3.87 | 7.95 | 4.00 | A | P |
| L13 | <1.00 | <1.00 | <2.00 | 1.00 | 7.96 | 2.30 | 6.83 | <2.00 | A | P |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Degenhardt, R.; Sobral Marques Souza, D.; Acordi Menezes, L.A.; de Melo Pereira, G.V.; Rodríguez-Lázaro, D.; Fongaro, G.; De Dea Lindner, J. Detection of Enteric Viruses and Core Microbiome Analysis in Artisanal Colonial Salami-Type Dry-Fermented Sausages from Santa Catarina, Brazil. Foods 2021, 10, 1957. https://doi.org/10.3390/foods10081957
Degenhardt R, Sobral Marques Souza D, Acordi Menezes LA, de Melo Pereira GV, Rodríguez-Lázaro D, Fongaro G, De Dea Lindner J. Detection of Enteric Viruses and Core Microbiome Analysis in Artisanal Colonial Salami-Type Dry-Fermented Sausages from Santa Catarina, Brazil. Foods. 2021; 10(8):1957. https://doi.org/10.3390/foods10081957
Chicago/Turabian StyleDegenhardt, Roberto, Doris Sobral Marques Souza, Leidiane A. Acordi Menezes, Gilberto Vinícius de Melo Pereira, David Rodríguez-Lázaro, Gislaine Fongaro, and Juliano De Dea Lindner. 2021. "Detection of Enteric Viruses and Core Microbiome Analysis in Artisanal Colonial Salami-Type Dry-Fermented Sausages from Santa Catarina, Brazil" Foods 10, no. 8: 1957. https://doi.org/10.3390/foods10081957
APA StyleDegenhardt, R., Sobral Marques Souza, D., Acordi Menezes, L. A., de Melo Pereira, G. V., Rodríguez-Lázaro, D., Fongaro, G., & De Dea Lindner, J. (2021). Detection of Enteric Viruses and Core Microbiome Analysis in Artisanal Colonial Salami-Type Dry-Fermented Sausages from Santa Catarina, Brazil. Foods, 10(8), 1957. https://doi.org/10.3390/foods10081957









