Some Important Metabolites Produced by Lactic Acid Bacteria Originated from Kimchi
Abstract
:1. Introduction
2. Important Metabolites Produced by Kimchi LAB
2.1. Bacteriocins
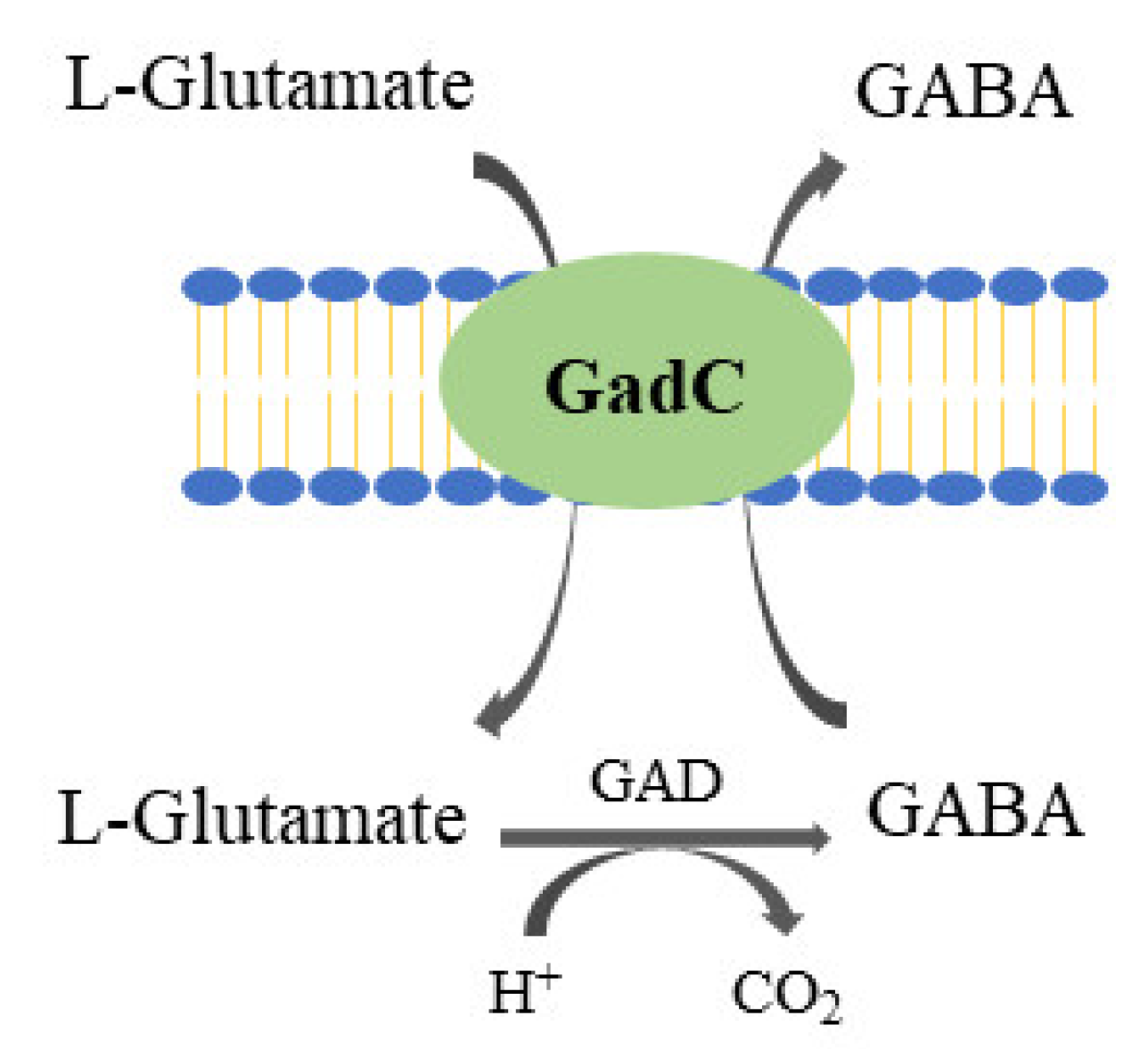
2.2. GABA (γ-Aminobutyric Acid)
2.3. Ornithine
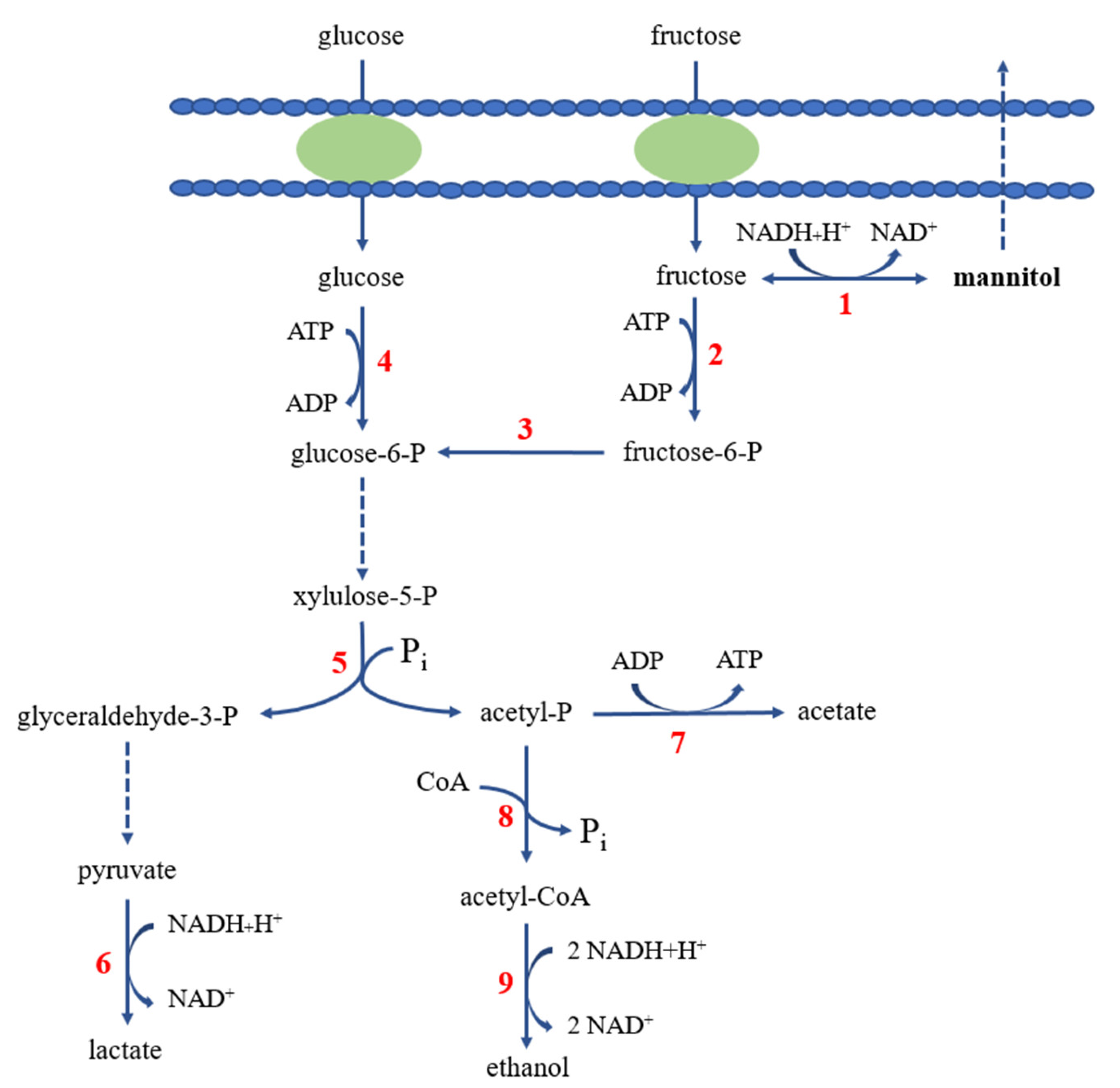
2.4. Mannitol
2.5. Exopolysaccharides
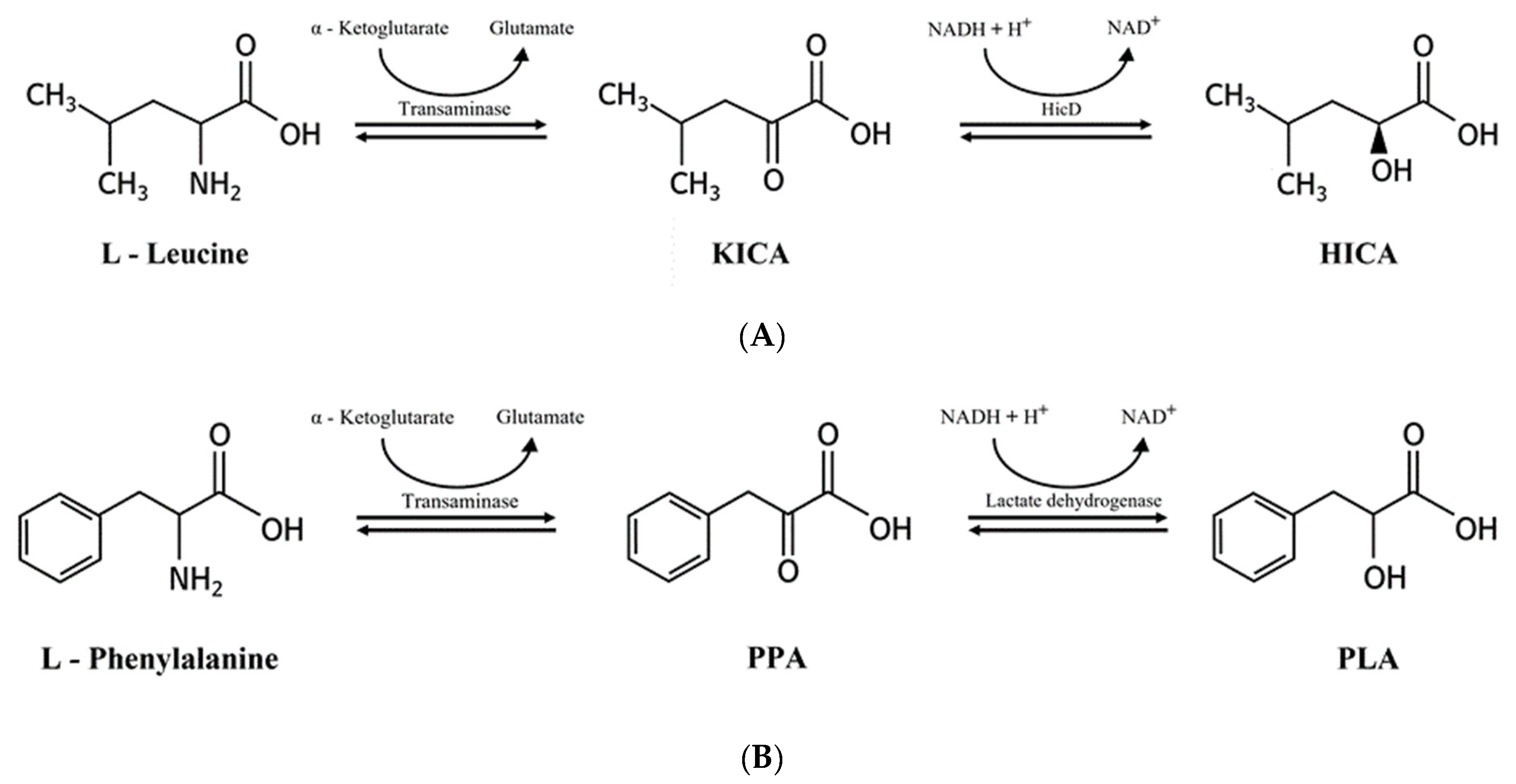
2.6. 2-Hydroxyisocaproic Acid and 3-Phenyllactic Acid
3. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Chang, J.Y.; Chang, H.C. Improvements in the quality and shelf life of kimchi by fermentation with the induced bacteriocin-producing strain, Leuconostoc citreum GJ7 as a starter. J. Food Sci. 2010, 75, M103–M110. [Google Scholar] [CrossRef]
- Lee, S.H.; Jung, J.Y.; Jeon, C.O. Source tracking and succession of kimchi lactic acid bacteria during fermentation. J. Food Sci. 2015, 80, M1871–M1877. [Google Scholar] [CrossRef] [PubMed]
- Park, K.Y.; Jeong, J.K.; Lee, Y.E.; Daily, J.W., III. Health benefits of kimchi (Korean fermented vegetables) as a probiotic food. J. Med. Food 2014, 17, 6–20. [Google Scholar] [CrossRef]
- Dimiti, E.; Cox, S.R.; Rossi, M.; Whelan, K. Fermented foods: Definition and characteristics, impact on the gut microbiota and effects on gastrointestinal health and disease. Nutrients 2019, 11, 1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Hutkins, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef]
- Park, E.J.; Chun, J.; Cha, C.J.; Park, W.S.; Jeon, C.O.; Bae, J.W. Bacterial community analysis during fermentation of ten representative kinds of kimchi with barcoded pyrosequencing. Food Microbiol. 2012, 30, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Lee, S.H.; Jeon, C.O. Kimchi microflora: History, current status, and perspectives for industrial kimchi production. Appl. Microbiol. Biotechnol. 2014, 98, 2385–2393. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.E.; Jang, J.Y.; Lee, J.H.; Park, H.W.; Choi, H.J.; Kim, T.W. Starter cultures for kimchi fermentation. J. Microbiol. Biotechnol. 2015, 25, 559–568. [Google Scholar] [CrossRef]
- Wuyts, S.; Beeck, W.V.; Allonsius, C.N.; van den Broek, M.F.L.; Lebeer, S. Applications of plant-based fermented foods and their microbes. Curr. Opin. Biotechnol. 2020, 61, 45–52. [Google Scholar] [CrossRef]
- Yu, A.; Leveau, J.H.J.; Marco, M.L. Abundance, diversity and plant-specific adaptations of plant-associated lactic acid bacteria. Environ. Microbiol. Rep. 2020, 12, 16–29. [Google Scholar] [CrossRef]
- Hatti-Kaul, R.; Chen, L.; Dishisa, T.; Enshasy, H.E. Lactic acid bacteria: From starter cultures to producers of chemicals. FEMS Microbiol. Lett. 2018, 365, fny213. [Google Scholar] [CrossRef] [Green Version]
- Hols, P.; Ledesma, G.L.; Gabant, P.; Mignolet, J. Mobilization of microbiota commensals and their bacteriocins for therapeutics. Trends Microbiol. 2019, 27, 690–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agriopoulou, S.; Stamatelopoulou, E.; Sachadyn-Krὁl, M.; Varzakas, T. Lactic acid bacteria as antibacterial agents to extend the shelf life of fresh and minimally processed fruits and vegetables: Quality and safety aspects. Microorganisms 2020, 8, 952. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, P.M.; Kuniyoshi, T.M.; Oliveira, R.P.S.; Hill, C.; Ross, R.P.; Cotter, P.D. Antimicrobials for food and feed; a bacteriocin perspective. Curr. Opin. Biotechnol. 2020, 61, 160–167. [Google Scholar] [CrossRef]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical applications of nisin. J. Appl. Microbiol. 2015, 120, 1449–1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.H.; Park, J.Y.; Jeong, S.J.; Kwon, G.H.; Lee, H.J.; Chang, H.C.; Kim, J.H. Characterization of Paraplantaricin C7, a Novel Bacteriocin Produced by Lactobacillus paraplantarum C7 Isolated from Kimchi. J. Microbiol. Biotechnol. 2007, 17, 287–296. Available online: https://www.koreascience.or.kr/article/JAKO200710912611025.pdf (accessed on 8 September 2021).
- Kim, H.T.; Park, J.Y.; Lee, G.G.; Kim, J.H. Isolation of a bacteriocin-producing Lactobacillus plantarum strain from kimchi. Food Sci. Biotechnol. 2003, 12, 166–170. [Google Scholar] [CrossRef]
- Hur, J.W.; Hyun, H.H.; Pyun, Y.R.; Kim, T.S.; Yeo, I.H.; Paik, H.D. Identification and partial characterization of lacticin BH5, a bacteriocin produced by Lactococcus lactis BH5 isolated from Kimchi. J. Food Prot. 2000, 63, 1707–1712. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Park, C.S.; Choi, N.S.; Yang, H.J.; Kim, C.Y.; Yoon, B.D.; Kim, M.S. Characteristics of bacteriocin produced by Lactococcus lactis ET45 isolated from kimchi. Korean J. Microbiol. 2011, 47, 74–80. [Google Scholar]
- Lee, N.K.; Jin Han, E.; Jun Han, K.; Paik, H.D. Antimicrobial effect of bacteriocin KU24 produced by Lactococcus lactis KU24 against methicillin-resistant Staphylococcus aureus: Characterization of bacteriocin KU24. J. Food Sci. 2013, 78, M465–M469. [Google Scholar] [CrossRef]
- Shin, J.Y.; Cheol, A. Characterization of Bacteriocin Production by Lactococcus lactis LAB3113 Isolated from Kimchi. J. Food Sci. Nutr. 1997, 2, 101–108. Available online: http://www.dbpia.co.kr/journal/articleDetail?nodeId=NODE00636627 (accessed on 8 September 2021).
- Lee, H.J.; Joo, Y.J.; Park, C.S.; Kim, S.H.; Hwang, I.K.; Ahn, J.S.; Mheen, T.I. Purification and characterization of a bacteriocin produced by Lactococcus lactis subsp. lactis H-559 isolated from kimchi. J. Biosci. Bioeng. 1999, 88, 153–159. [Google Scholar] [CrossRef]
- Kwak, G.S.; Kim, S.K.; Jun, H.K. Purification and characterization of bacteriocin J105 produced by Lactococcus latis subsp. lactis J105 isolated from kimchi. J. Microbiol. Biotechnol. 2001, 11, 275–280. [Google Scholar]
- Kim, S.K.; Lee, E.J.; Park, K.Y.; Jun, H.K. Bacteriocin produced by Lactobacillus curvatus SE1 isolated from kimchi. J. Microbiol. Biotechnol. 1998, 8, 588–594. [Google Scholar]
- Ahn, J.E.; Kim, J.K.; Lee, H.R.; Eom, H.J.; Han, N.S. Isolation and characterization of a bacteriocin-producing Lactobacillus sakei B16 from kimchi. J. Korean Soc. Food Sci. Nutr. 2012, 41, 721–726. [Google Scholar] [CrossRef] [Green Version]
- Moon, S.H.; Kim, C.R.; Chang, H.C. Heterofermentative lactic acid bacteria as a starter culture to control kimchi fermentation. LWT-Food Sci. Technol. 2018, 88, 181–188. [Google Scholar] [CrossRef]
- Chang, J.Y.; Lee, H.J.; Chang, H.C. Identification of the agent from Lactobacillus plantarum KFRI464 that enhances bacteriocin production by Leuconostoc citreum GJ7. J. Appl. Microbiol. 2007, 103, 2504–2515. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.C.; Oh, J.K.; Kim, S.H.; Oh, S.; Kang, D.K. Isolation and characterization of an anti-listerial bacteriocin from Leuconostoc lactis SD501. Korean J. Food Sci. Anim. Resour. 2018, 38, 1008–1018. [Google Scholar] [CrossRef] [Green Version]
- Shin, M.S.; Han, S.K.; Ryu, J.S.; Kim, K.S.; Lee, W.K. Isolation and partial characterization of a bacteriocin produced by Pediococcus pentosaceus K23-2 isolated from kimchi. J. Appl. Microbiol. 2008, 105, 331–339. [Google Scholar] [CrossRef]
- Jang, S.; Lee, J.; Jung, U.; Choi, H.S.; Suh, H.J. Identification of an anti-listerial domain from Pediococcus pentosaceus T1 derived from kimchi, a traditional fermented vegetable. Food Control 2014, 43, 42–48. [Google Scholar] [CrossRef]
- Miller, E.R.; Kearns, P.J.; Niccum, B.A.; Schwartz, J.Ó.; Ornstein, A.; Wolfe, B.E. Establishment limitation constrains the abundance of lactic acid bacteria in the Napa cabbage phyllosphere. Appl. Environ. Microbiol. 2019, 85, e00269-19. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.W.; Kim, G.S.; Baek, A.H.; Hwang, H.S.; Kwon, D.Y.; Kim, S.G.; Lee, S.Y. Isolation and characterization of kimchi starters Leuconostoc mesenteroides PBio03 and Leuconostoc mesenteroides PBio04 for manufacture of commercial kimchi. J. Microbiol. Biotechnol. 2020, 30, 1060–1066. [Google Scholar] [CrossRef]
- Walsh, C.J.; Guinane, C.M.; Hill, C.; Ross, R.P.; O’Toole, P.W.; Cotter, P.D. In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the human microbiome project’s reference genome database. BMC Microbiol. 2015, 15, 183. [Google Scholar] [CrossRef] [Green Version]
- Rea, M.C.; Dobson, A.; O’Sullivan, O.; Crispie, F.; Fouhy, F.; Cotter, P.D.; Shanahan, F.; Kiely, B.; Hill, C.; Ross, R.P. Effect of broad and narrow-spectrum antimicrobials on clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl. Acad. Sci. USA 2011, 108, 4639–4644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Małaczewska, J.; Kaczorek-Łukowska, E. Nisin—A lantibiotic with immunomodulatory properties: A review. Peptides 2021, 137, 170479. [Google Scholar] [CrossRef] [PubMed]
- Kamarajan, P.; Ateia, I.; Shin, J.M.; Fenno, J.C.; Le, C.; Zhan, L.; Chang, A.; Darveau, R.; Kapila, Y.L. Periodontal pathogens promote cancer aggressivity via TLR/MyD88 triggered activation of Integrin/FAK signaling that is therapeutically reversible by a probiotic bacteriocin. PLoS Pathog. 2020, 16, 1–27. [Google Scholar] [CrossRef]
- Field, D.; Cotter, P.D.; Ross, R.P.; Hil, C. Bioengineering of the model lantibiotic nisin. Bioengineered 2015, 6, 187–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Miao, K.; Niyaphorn, S.; Qu, X. Production of gamma-aminobutyric acid from lactic acid bacteria: A systematic review. Int. J. Mol. Sci. 2020, 21, 995. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.; Wei, L.; Liu, J. Biotechnological advances and perspectives of gamma-aminobutyric acid production. World J. Microbiol. Biotechnol. 2017, 33, 64. [Google Scholar] [CrossRef]
- Ngo, D.H.; Vo, T.S. An updated review on pharmaceutical properties of gamma-aminobutyric acid. Molecules 2019, 24, 2678. [Google Scholar] [CrossRef] [Green Version]
- Yogeswara, I.B.A.; Maneerat, S.; Haltrich, D. Glutamate decarboxylase from lactic acid bacteria—A key enzyme in GABA synthesis. Microorganisms 2020, 8, 1923. [Google Scholar] [CrossRef]
- Sarasa, S.B.; Mahendran, R.; Muthusamy, G.; Thankappan, B.; Selta, D.R.F.; Angayarkanni, J. A brief review on the non-protein amino acid, gamma-amino butyric acid (GABA): Its production and role in microbes. Curr. Microbiol. 2020, 77, 534–544. [Google Scholar] [CrossRef]
- Kim, J.; Bang, J.; Beuchat, L.R.; Kim, H.; Ryu, J.H. Controlled fermentation of kimchi using naturally occurring antimicrobial agents. Food Microbiol. 2012, 32, 20–31. [Google Scholar] [CrossRef]
- Lee, K.W.; Shim, J.M.; Yao, Z.; Kim, J.A.; Kim, H.J.; Kim, J.H. Characterization of a glutamate decarboxylase (GAD) from Enterococcus avium M5 isolated from jeotgal, a korean fermented seafood. J. Microbiol. Biotechnol. 2017, 27, 1216–1222. [Google Scholar] [CrossRef]
- Lim, H.S.; Cha, I.T.; Lee, H.; Seo, M.J. Optimization of γ-aminobutyric acid production by Enterococcus faecium JK29 isolated from a traditional fermented foods. Microbiol. Biotechnol. Lett. 2016, 44, 26–33. [Google Scholar] [CrossRef]
- Park, S.Y.; Lee, J.W.; Lim, S.D. The probiotic characteristics and GABA production of Lactobacillus plantarum K154 isolated from kimchi. Food Sci. Biotechnol. 2014, 23, 1951–1957. [Google Scholar] [CrossRef]
- Lu, X.; Chen, Z.; Gu, Z.; Han, Y. Isolation of γ-aminobutyric acid-producing bacteria and optimization of fermentative medium. Biochem. Eng. J. 2008, 41, 48–52. [Google Scholar] [CrossRef]
- Sa, H.D.; Park, J.Y.; Jeong, S.J.; Lee, K.W.; Kim, J.H. Characterization of glutamate decarboxylase (GAD) from Lactobacillus sakei A156 isolated from jeot-gal. J. Microbiol. Biotechnol. 2015, 25, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.J.; Oh, S.H. γ-aminobutyric acid production and glutamate decarboxylase activity of Lactobacillus sakei OPK2-59 isolated from kimchi. Korean J. Microbiol. Biotechnol. 2011, 47, 316–322. [Google Scholar]
- Cho, S.Y.; Park, M.J.; Kim, K.M.; Ryu, J.H.; Park, H.J. Production of high γ-aminobutyric acid (GABA) sour kimchi using lactic acid bacteria isolated from mukeunjee kimchi. Food Sci. Biotechnol. 2011, 20, 403–408. [Google Scholar] [CrossRef]
- Cho, Y.R.; Chang, J.Y.; Chang, H.C. Production of γ-aminobutyric acid (GABA) by Lactobacillus buchneri isolated from kimchi and its neuroprotective effect on neuronal cells. J. Microbiol. Biotechnol. 2007, 17, 104–109. [Google Scholar]
- Seo, M.J.; Lee, J.Y.; Nam, Y.D.; Lee, S.Y.; Park, S.L.; Yi, S.H.; Lim, S.I. Production of γ-aminobutyric acid by Lactobacillus brevis 340G isolated from kimchi and its application to skim milk. Food Eng. Prog. 2013, 17, 418–423. [Google Scholar] [CrossRef]
- Seo, M.J.; Nam, Y.D.; Lee, S.Y.; Park, S.L.; Yi, S.H.; Lim, S.I. Expression and characterization of a glutamate decarboxylase from Lactobacillus brevis 877G producing γ-aminobutyric acid. Biosci. Biotechnol. Biochem. 2013, 77, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Park, J.Y.; Kim, J.H. Characterization of the recombinant glutamate decarboxylase of Lactobacillus brevis G144 isolated from galchi jeotgal, a Korean salted and fermented seafood. Microbiol. Biotechnol. Lett. 2021, 49, 9–17. [Google Scholar] [CrossRef]
- Lim, H.S.; Cha, I.T.; Roh, S.W.; Shin, H.H.; Seo, M.J. Enhanced production of gamma-aminobutyric acid by optimizing culture conditions of Lactobacillus brevis HYE1 isolated from kimchi, a Korean fermented food. J. Microbiol. Biotechnol. 2017, 27, 450–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binh, T.T.T.; Ju, W.T.; Jung, W.J.; Park, R.D. Optimization of γ-amino butyric acid production in a newly isolated Lactobacillus brevis. Biotechnol. Lett. 2014, 36, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Shah, N.P. Gas release-based prescreening combined with reversed-phase HPLC quantitation for efficient selection of high-γ-aminobutyric acid (GABA)-producing lactic acid bacteria. J. Dairy Sci. 2015, 98, 790–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.J.; Yao, Z.; Meng, Y.; Le, H.G.; Jeon, H.S.; Yoo, J.Y.; Kim, J.H. Isolation of γ-aminobutyric acid producing Lactobacillus brevis T118 from Sun-Tae Jeotgal and its glutamate decarboxylase gene cloning. J. Agric. Life Sci. 2020, 54, 85–92. [Google Scholar] [CrossRef]
- Park, J.Y.; Jeong, S.J.; Kim, J.H. Characterization of a glutamate decarboxylase (GAD) gene from Lactobacillus zymae. Biotechnol. Lett. 2014, 36, 1791–1799. [Google Scholar] [CrossRef]
- Jung, M.Y.; Kim, T.W.; Lee, C.; Kim, J.Y.; Song, H.S.; Kim, Y.B.; Ahn, S.W.; Kim, J.S.; Roh, S.W.; Lee, S.H. Role of jeotgal, a korean traditional fermented fish sauce, in microbial dynamics and metabolite profiles during kimchi fermentation. Food Chem. 2018, 265, 135–143. [Google Scholar] [CrossRef]
- Lee, K.W.; Shim, J.M.; Yao, Z.; Kim, J.A.; Kim, J.H. Properties of kimchi fermented with GABA-producing lactic acid bacteria as a starter. J. Microbiol. Biotechnol. 2018, 28, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Fan, E.; Huang, J.; Hu, S.; Mei, L.; Yu, K. Cloning, sequencing and expression of a glutamate decarboxylase gene from the GABA-producing strain Lactobacillus brevis CGMCC 1306. Ann. Microbiol. 2012, 62, 689–698. [Google Scholar] [CrossRef]
- Gong, L.; Ren, C.; Xu, Y. Deciphering the crucial roles of transcriptional regulator GadR on gamma-aminobutyric acid production and acid resistance in Lactobacillus brevis. Microb. Cell Factories 2019, 18, 108. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.; Lee, J.H. Characterization of arginine catabolism by lactic acid bacteria isolated from kimchi. Molecules 2018, 23, 3049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, H.; Li, Y.; Yun, H.; Zhang, T.; Huang, Y.; Zhou, J.; Yan, H.; Wei, J.; Liu, Y.; Zhang, Z.; et al. Lactobacillus maintains healthy gut mucosa by producing L-ornithine. Commun. Biol. 2019, 2, 171. [Google Scholar] [CrossRef] [Green Version]
- Majsnerowska, M.; Noens, E.E.; Lolkema, J.S. Arginine and citrulline catabolic pathways encoded by the arc gene cluster of Lactobacillus brevis ATCC 367. J. Bacteriol. 2018, 200, e00182-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noens, E.E.; Lolkema, J.S. Convergent evolution of the arginine deiminase pathway: The ArcD and ArcE arginine/ornithine exchangers. Microbiologyopen 2017, 6, e00412. [Google Scholar] [CrossRef]
- Su, M.S.; Schlicht, S.; Gänzle, M.Z. Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb. Cell Factories 2011, 10 (Suppl. S1), S8. [Google Scholar] [CrossRef] [Green Version]
- Kang, B.K.; Cho, M.S.; Ahn, T.Y.; Lee, E.S.; Park, D.S. The influence of red pepper powder on the density of Weissella koreensis during kimchi fermentation. Sci. Rep. 2015, 5, 15445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, S.; Lee, J.H. Red pepper powder is an essential factor for ornithine production in kimchi fermentation. LWT-Food Sci. Technol. 2021, 137, 110434. [Google Scholar] [CrossRef]
- Park, J.E.; Oh, S.H.; Cha, Y.S. Lactobacillus brevis OPK-3 from kimchi prevents obesity and modulates the expression of adipogenic and pro-inflammatory genes in adipose tissue of diet-induced obese mice. Nutrients 2020, 12, 604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mun, S.Y.; Moon, S.H.; Chang, H.C. Characterization of high-ornithine-producing Weissella koreensis DB1 isolated from kimchi and its application in rice bran fermentation as a starter culture. Foods 2020, 9, 1545. [Google Scholar] [CrossRef]
- Yu, J.J.; Park, H.J.; Kim, S.G.; Oh, S.H. Isolation, identification, and characterization of Weissella strains with high ornithine producing capacity from kimchi. Korean J. Microbiol. 2009, 45, 339–345. [Google Scholar]
- Mun, S.Y.; Chang, H.C. Characterization of Weissella koreensis SK isolated from kimchi fermented at low temperature (around 0 °C) based on complete genome sequence and corresponding phenotype. Microorganisms 2020, 8, 1147. [Google Scholar] [CrossRef]
- Park, J.A.; Tirupathi Pichiah, P.B.; Yu, J.J.; Oh, S.H.; Daily, J.W., III; Cha, Y.S. Anti-obesity effect of kimchi fermented with Weissella koreensis OK 1-6 as starter in high-fat diet-induced obese C57BL/6J mice. J. Appl. Microbiol. 2012, 113, 1507–1516. [Google Scholar] [CrossRef]
- Wisselink, H.W.; Weusthuis, R.A.; Eggink, G.; Hugenholtz, J.; Grobben, G.J. Mannitol production by lactic acid bacteria: A review. Int. Dairy J. 2002, 12, 151–161. [Google Scholar] [CrossRef]
- Park, Y.C.; Oh, E.J.; Jo, J.H.; Jin, Y.S.; Seo, J.H. Recent advances in biological production of sugar alcohols. Curr. Opin. Biotechnol. 2016, 37, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Rice, T.; Sahin, A.W.; Lynch, K.M.; Arendt, E.K.; Coffey, A. Isolation, characterisation and exploitation of lactic acid bacteria capable of efficient conversion of sugars to mannitol. Int. J. Food Microbiol. 2020, 321, 108546. [Google Scholar] [CrossRef]
- Helanto, M.; Aarnikunnas, J.; von Weymarn, N.; Airaksinen, U.; Palva, A.; Leisola, M. Improved mannitol production by a random mutant of Leuconostoc pseudomesenteroides. J. Biotechnol. 2005, 116, 283–294. [Google Scholar] [CrossRef]
- Wisselink, H.W.; Moers, A.P.H.A.; Mars, A.E.; Hoefnagel, M.H.N.; de Vos, W.M.; Hugenholtz, J. Overproduction of heterologous mannitol 1-phosphatase: A key factor for engineering mannitol production by Lactococcus lactis. Appl. Environ. Microbiol. 2005, 71, 1507–1514. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Gu, L.; Cheng, C.; Ma, J.; Xin, F.; Liu, J.; Wu, H.; Jiang, M. Recent advances in microbial production of mannitol: Utilization of low-cost substrates, strain development and regulation strategies. World J. Microbiol. Biotechnol. 2018, 34, 49. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.C.; Racine, F.M. Biotechnological production of mannitol and its application. Appl. Microbial. Biotechnol. 2011, 89, 879–891. [Google Scholar] [CrossRef]
- Dai, Y.; Meng, Q.; Mu, W.; Zhang, T. Recent advances in the applications and biotechnological production of mannitol. J. Funct. Foods 2017, 36, 404–409. [Google Scholar] [CrossRef]
- Martău, G.A.; Coman, V.; Vodnar, D.C. Recent advances in the biotechnological production of erythritol and mannitol. Crit. Rev. Biotechnol. 2020, 40, 608–622. [Google Scholar] [CrossRef] [PubMed]
- Otgonbayar, G.E.; Eom, H.J.; Kim, B.S.; Ko, J.H.; Han, N.S. Mannitol production by Leuconostoc citrum KACC91348P isolated from kimchi. J. Microbiol. Biotechnol. 2011, 21, 968–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, J.Y.; Lee, S.H.; Lee, H.J.; Seo, H.Y.; Park, W.S.; Jeon, C.O. Effects of Leuconostoc mesenteroides starter cultures on microbial communities and metabolites during kimchi fermentation. Int. J. Food Microbiol. 2012, 153, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Lee, H.W.; Lee, M.E.; Roh, S.W.; Kim, T.W. Mixed starter of Lactococcus lactis and Leuconostoc citreum for extending kimchi shelf-life. J. Microbiol. Biotechnol. 2019, 57, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Xing, H.; Yang, Y.; Jiang, H.; Zhou, Z.; Han, Y. Optimization, purification and structural characterization of a dextran produced by L. mesenteroides isolated from Chinese sauerkraut. Carbohydr. Polym. 2017, 174, 409–416. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Hols, P.; Bernard, E.; Rolain, T.; Zhou, M.; Siezen, R.J.; Bron, P.A. The extracellular biology of the lactobacilli. FEMS Microbiol. Rev. 2010, 34, 199–230. [Google Scholar] [CrossRef]
- Chen, F.; Huang, G.; Huang, H. Preparation and application of dextran and its derivatives as carriers. Int. J. Biol. Macromol. 2020, 145, 827–834. [Google Scholar] [CrossRef]
- Daba, G.M.; Elnahas, M.O.; Elkhateeb, W.A. Contribution of exopolysaccharides from lactic acid bacteria as biotechnological tools in food, pharmaceutical, and medical applications. Int. J. Biolog. Macromol. 2021, 173, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Majumder, A.; Goyal, A. Potentials of exopolysaccharides from lactic acid bacteria. Indian J. Microbiol. 2012, 52, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Ramos, A.; Nácher-Vázquez, M.; Notararigo, S.; López, P.; Mohedano, M.L. Current and future applications of bacterial extracellular polysaccharides. In Probiotics, Prebiotics, and Synbiotics; Preedy, V.R., Watson, R.R., Eds.; Elsevier: Oxford, UK, 2015; pp. 329–344. [Google Scholar] [CrossRef]
- Angelin, J.; Kavitha, M. Exopolysaccharides from probiotic bacteria and their health potential. Int. J. Biolog. Macromol. 2020, 162, 853–865. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Y.; Ye, L.; Wang, C. The anti-cancer effects and mechanisms of lactic acid bacteria exopolysaccharides in vitro: A review. Carbohydr. Polym. 2021, 253, 117308. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lee, G.; Thanh, H.D.; Kim, J.H.; Konkit, M.; Yoon, S.; Park, M.; Yang, S.; Park, E.; Kim, W. Exopolysaccharide from Lactobacillus plantarum LRCC5310 offers protection against rotavirus-induced diarrhea and regulates inflammatory response. J. Dairy Sci. 2018, 101, 5702–5712. [Google Scholar] [CrossRef] [PubMed]
- Kook, S.Y.; Lee, Y.; Jeong, E.C.; Kim, S. Immunomodulatory effects of exopolysaccharides produced by Bacillus licheniformis and Leuconostoc mesenteroides isolated from Korean kimchi. J. Funct. Foods 2019, 54, 211–219. [Google Scholar] [CrossRef]
- Zarour, K.; Llamas, M.G.; Prieto, A.; Rúas-Madiedo, P.; Dueñas, M.T.; de Palencia, P.F.; Aznar, R.; Kihal, M.; López, P. Rheology and bioactivity of high molecular weight dextrans synthesised by lactic acid bacteria. Carbohydr. Polym. 2017, 174, 646–657. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.S.; Jang, H.J.; Lee, N.K.; Paik, H.D. Evaluation of the probiotic characteristics and prophylactic potential of Weissella cibaria strains isolated from kimchi. LWT-Food Sci. Technol. 2019, 112, 108229. [Google Scholar] [CrossRef]
- Yadav, R.; Singh, P.K.; Shukla, P. Metabolic engineering for probiotics and their genome-wide expression profiling. Curr. Protein Pept. Sci. 2018, 19, 68–74. [Google Scholar] [CrossRef]
- Li, N.; Wang, Y.; Zhu, P.; Liu, Z.; Guo, B.; Ren, J. Improvement of exopolysaccharide production in Lactobacillus casei LC2W by overexpression of NADH oxidase gene. Microbiol. Res. 2015, 171, 73–77. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, G.; Ahmad, T.; Kaur, B.; Hakeem, K.R. Tailoring cellular metabolism in lactic acid bacteria through metabolic engineering. J. Microbiol. Methods 2020, 170, 105862. [Google Scholar] [CrossRef]
- Axel, C.; Brosnan, B.; Zannini, E.; Peyer, L.C.; Furey, A.; Coffey, A.; Arendt, E. Antifungal activities of three different Lactobacillus species and their production of antifungal carboxylic acids in wheat sourdough. Appl. Microbiol. Biotechnol. 2016, 100, 1701–1711. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Li, H.; Song, D.; Zhang, L.; Zhao, X.; Xu, X. Genome, transcriptome and fermentation analyses of Lactobacillus plantarum LY-78 provide new insights into the mechanism of phenyllactate biosynthesis in lactic acid bacteria. Biochem. Biophys. Res. Commun. 2019, 519, 351–357. [Google Scholar] [CrossRef]
- Park, B.; Hwang, H.; Chang, J.; Hong, S.W.; Lee, S.H.; Jung, M.Y.; Sohn, S.O.; Park, H.W.; Lee, J.H. Identification of 2-hydroxyisocaproic acid production in lactic acid bacteria and evaluation of microbial dynamics during kimchi ripening. Sci. Rep. 2017, 7, 10904. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, N.; Gaänzle, M.G.; Vogel, R.F. Influence of peptide supply and cosubstrates on phenylalanine metabolism of Lactobacillus sanfranciscensis DSM20451T and Lactobacillus plantarum TMW1.468. J. Agric. Food Chem. 2006, 54, 3832–3839. [Google Scholar] [CrossRef]
- Zheng, R.; Zhao, T.; Hung, Y.C.; Adhikari, K. Evaluation of bactericidal effects of phenyllactic acid on Escherichia coli O157:H7 and Salmonella Typhimurium on beef meat. J. Food Prot. 2019, 82, 2016–2022. [Google Scholar] [CrossRef] [PubMed]
- Mero, A.A.; Ojala, T.; Hulmi, J.J.; Puurtinen, R.; Kalira, T.A.M.; Seppälä, T. Effects of alpha-hydroxy-isocaproic acid on body composition, DOMS and performance in athletes. J. Int. Soc. Sports Nutr. 2010, 7, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixera, F.; Matias, C.N.; Monteiro, C.P.; Valamatos, M.J.; Reis, J.F.; Tavares, F.; Batista, A.; Domingos, C.; Alves, F.; Sardinha, L.B.; et al. Leucine metabolites do not enhance training-induced performance or muscle thickness. Med. Sci. Sports Exerc. 2019, 51, 56–64. [Google Scholar] [CrossRef]
- Fujita, T.; Nguyen, H.D.; Ito, T.; Zhou, S.; Osada, L.; Tateyama, S.; Kaneko, T.; Takaya, N. Microbial monomers custom-synthesized to build true bio-derived aromatic polymers. Appl. Microbiol. Biotechnol. 2013, 97, 8887–8894. [Google Scholar] [CrossRef] [PubMed]
- Ilavenil, S.; Kim, D.H.; Arasu, M.V.; Srigopalram, S.; Sivanesan, R.; Choi, K.C. Phenyllactic acid from Lactobacillus plantarum promotes adipogenic activity in 3T3-L1 adipocyte via up-regulation of PPAR-γ2. Molecules 2015, 20, 15359–15373. [Google Scholar] [CrossRef] [Green Version]
- Peters, A.; Krumbholz, P.; Jäger, E.; Heintz-Buschart, A.; Cakir, M.V.; Rothemund, S.; Gaudl, A.; Ceglarek, U.; Schőneberg, T.; Stäubert, C. Metabolites of lactic acid bacteria present in fermented foods are highly potent agonists of human hydroxycarboxylic acid receptor 3. PLoS Genet. 2019, 15. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.; Lee, J.H. Evaluation of metabolites derived from lactic acid bacteria isolated from kimchi. In Chemistry of Korean Foods and Beverages; Do, C.H., Rimando, A.M., Kim, Y., Eds.; ACS Publications: Washington, DC, USA, 2019; Volume 1303, pp. 3–10. [Google Scholar] [CrossRef] [Green Version]
- Sakko, M.; Tjäderhane, L.; Sorsa, T.; Hietala, P.; Rautemaa, R. 2-Hydroxyisocaproic acid is bactericidal in human dental root canals ex vivo. Int. Endod. J. 2017, 50, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Nieminen, M.T.; Novak-Frazer, L.; Rautemaa, V.; Rajendran, R.; Sorsa, T.; Ramage, G.; Bowyer, P.; Rautemaa, R. A novel antifungal is active against Candida albicans biofilms and inhibits mutagenic acetaldehyde production in vitro. PLoS ONE 2014, 9, e97864. [Google Scholar] [CrossRef]
- Sakko, M.; Moore, C.; Novak-Frazer, L.; Rautemaa, V.; Sorsa, T.; Hietala, P.; Järvinen, A.; Bowyer, P.; Tjäderhane, L.; Rautemaa, R. 2-Hydroxyisocaproic acid is fungicidal for Candida and Aspergillus species. Mycoses 2014, 57, 214. [Google Scholar] [CrossRef] [PubMed]
- Filannino, P.; Cagno, R.D.; Gobbetti, M. Metabolic and functional paths of lactic acid bacteria in plant foods: Get out of the labyrinth. Curr. Opin. Biotechnol. 2018, 49, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Kim, B.; Ban, J.; Lee, J.; Kim, B.J.; Choi, B.S.; Hwang, S.; Ahn, K.; Kim, J. A randomized trial of Lactobacillus plantarum CJLP133 for the treatment of atopic dermatitis. Pediatr. Allergy Immunol. 2012, 23, 667–673. [Google Scholar] [CrossRef] [PubMed]

| Strain | pH Stability | Temperature (°C) Stability | Molecular Size (kDa) | Reference |
|---|---|---|---|---|
| Lactiplantibacillus paraplantarum C7 | 2 to 8 a, 9 b, 10 c | 100 °C, 20 min, 121 °C, 10 min b, 100 °C, 30 min, 121 °C, 30 min c | 3.8 | [16] |
| Lactiplantibacillus plantarum J9 | 3 to 10 a | 100 °C, 60 min, 121 °C, 15 min a | less than 6.5 | [17] |
| Lactococcus lactis BH5 | 2 to 9 a | 90 °C, 30 min a, 100 °C, 30 min d | 3.7 | [18] |
| Lactococcus lactis ET45 | 3 to 5 a, 7 to 11 c | 121 °C, 60 min a | 4.5 | [19] |
| Lactococcus lactis KU24 | 3 to 7 a, 8 to 9 c | 100 °C, 30 min a, 121 °C, 15 min c | 6.5 | [20] |
| Lactococcus lactis LAB3113 | 2 to 10 a | 100 °C, 30 min, 121 °C, 20 min d | 10.5 | [21] |
| Lactococcus lactis subsp. lactis H-559 | 2 to 11 e | 100 °C, 10 min a, 100 °C, 30 min, 121 °C, 10 min c, 121 °C, 20 min d | 3.3 | [22] |
| Lactococcus lactis subsp. lactis J105 | 3 a, 2, 4 b, 5 c, 6 to 9 d | 4 °C, 24 h, 100 °C, 1 h, 110 °C, 10 min, 121 °C, 15 min e | 3.4 | [23] |
| Latilactobacillus curvatus SE1 | 2 to 11 e | 100 °C, 60 min e | 14 | [24] |
| Latilactobacillus sakei B16 | 2 to 9 e | 100 °C, 30 min, 121 °C, 15 min e | ND * | [25] |
| Leuconostoc citreum C2 | 3 to 4 a, 5 to 7 d | 50 °C, 24 h a, 70 °C, 24 h d | ND | [26] |
| Leuconostoc citreum GJ7 | 2.5 to 9.5 a | 70 °C, 24 h, 100 °C, 30 min, 121 °C, 15 min a | 3.5 | [27] |
| Leuconostoc citreum GR1 | 3 to 4 a, 5 to 7 d | 70 °C, 24 h, 100 °C, 30 min, 121 °C, 15 min a | ND | [26] |
| Leuconostoc lactis SD501 | 2 to 10 e | 121 °C, 15 min e | 7 | [28] |
| Pediococcus pentosaceus K23-2 | 2 to 7 a, 8 c | 95 °C, 30 min, 121 °C, 15 min a | 5 | [29] |
| Pediococcus pentosaceus T1 | 4 to 8 e | 110 °C, 20 min e | 23 | [30] |
| Strain (Microorganisms) | GABA Content | MSG Concentration | Reference |
|---|---|---|---|
| Enterococcus avium M5 a | 18.47 mg/mL | MRS + 3% MSG | [44] |
| Enterococcus faecium JK29 | 14.86 mM | MRS + 0.5% MSG | [45] |
| Lactiplantibacillus plantarum K154 | 201.78 µg/mL | MRS + 3% MSG | [46] |
| Lactococcus lactis subsp. lactis B | 3.68 g/L | MRS + 1% MSG | [47] |
| Latilactobacillus sakei A156 a | 15.81 mg/mL | MRS + 3% MSG | [48] |
| Latilactobacillus sakei OPK 2-59 | 58.88 mM | MRS + 1% MSG | [49] |
| Lentilactobacillus buchneri | 5.83 mg /mL | MRS + 50 mM glutamate | [50] |
| Lentilactobacillus buchneri MS | 251 mM | MRS + 5% MSG | [51] |
| Levilactobacillus brevis 340G | 68.77 mM | MRS + 3% MSG | [52] |
| Levilactobacillus brevis 877G | 18.51 mmol/L | MRS + 1% MSG | [53] |
| Levilactobacillus brevis G144 a | 14.58 mM | MRS + 3% MSG | [54] |
| Levilactobacillus brevis HYE1 | 18.76 mM | MRS + 2.38% MSG | [55] |
| Levilactobacillus brevis K203 | 44.4 g/L | MRS + 6% L-glutamate | [56] |
| Levilactobacillus brevis NPS-QW-145 | 25.83 g/L | MRS + 7% MSG | [57] |
| Levilactobacillus brevis NPS-QW-267 | 24.99 g/L | MRS + 7% MSG | [57] |
| Levilactobacillus brevis T118 a | ND b | MRS + 3% MSG | [58] |
| Levilactobacillus zymae GU240 | 16.94 mg/mL | MRS + 3% MSG | [48,59] |
| Strain | MSG Concentration | Operon Structure | GAD Size (aa) | Optimal pH | Optimal Temp. (°C) | Km (mM) | Vmax | Reference |
|---|---|---|---|---|---|---|---|---|
| Enterococcus avium M5 a | MRS + 3% MSG | gadCB | 466 | 4.5 | 55 | 3.26 | 0.012 mM/min | [44] |
| Latilactobacillus sakei A156 a | MRS + 3% MSG | gadCB | 479 | 5.0 | 55 | 16.0 | 0.011 mM/min | [48] |
| Latilactobacillus sakei OPK 2-59 | MRS + 1% MSG | ND b | ND | 5.0 c | 30 c | ND | ND | [49] |
| Levilactobacillus brevis 877G | MRS + 1% MSG | ND | 468 | 5.2 | 45 | 3.6 | 0.06 mM/min | [53] |
| Levilactobacillus brevis CGMCC 1306 | 0.17% MSGc | ND | 468 | 4.8 | 48 | 10.26 | 8.86 U/mg | [62] |
| Levilactobacillus brevis G144 a | MRS + 3% MSG | gadCB | 479 | 5.0 | 40 | 8.6 | 0.01 mM/min | [54] |
| Levilactobacillus zymae GU240 | MRS + 3% MSG | gadCB | 479 | 4.5 | 41 | 1.7 | 0.01 mM/min | [59] |
| Strain | Ornithine Yield | Arginine Concentration | Reference |
|---|---|---|---|
| Levilactobacillus brevis OPK-3 | ND * | MRS + 4% arginine | [71] |
| Weissella koreensis DB1 | 43.07 g/kg | MRS + 3% arginine | [72] |
| Weissella koreensis OK1-4 | 27.01 mg/L/h | MRS + 1% arginine | [73] |
| Weissella koreensis OK1-6 | 31.41 mg/L/h | MRS + 1% arginine | [73] |
| Weissella koreensis SK | 7.17 g/L | MRS + 1% arginine | [74] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-J.; Jeon, H.-S.; Yoo, J.-Y.; Kim, J.-H. Some Important Metabolites Produced by Lactic Acid Bacteria Originated from Kimchi. Foods 2021, 10, 2148. https://doi.org/10.3390/foods10092148
Lee S-J, Jeon H-S, Yoo J-Y, Kim J-H. Some Important Metabolites Produced by Lactic Acid Bacteria Originated from Kimchi. Foods. 2021; 10(9):2148. https://doi.org/10.3390/foods10092148
Chicago/Turabian StyleLee, Se-Jin, Hye-Sung Jeon, Ji-Yeon Yoo, and Jeong-Hwan Kim. 2021. "Some Important Metabolites Produced by Lactic Acid Bacteria Originated from Kimchi" Foods 10, no. 9: 2148. https://doi.org/10.3390/foods10092148
APA StyleLee, S.-J., Jeon, H.-S., Yoo, J.-Y., & Kim, J.-H. (2021). Some Important Metabolites Produced by Lactic Acid Bacteria Originated from Kimchi. Foods, 10(9), 2148. https://doi.org/10.3390/foods10092148





