Non-Targeted LC-MS Metabolomics Approach towards an Authentication of the Geographical Origin of Grain Maize (Zea mays L.) Samples †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Chemicals
2.2. Grain Maize Samples
2.3. Sample Treatment
2.4. LC-MS(/MS)-Analysis
2.5. Data Processing
2.6. Multivariate Data Analysis and Classification
3. Results and Discussion
3.1. General Evaluation of the Analytical Method
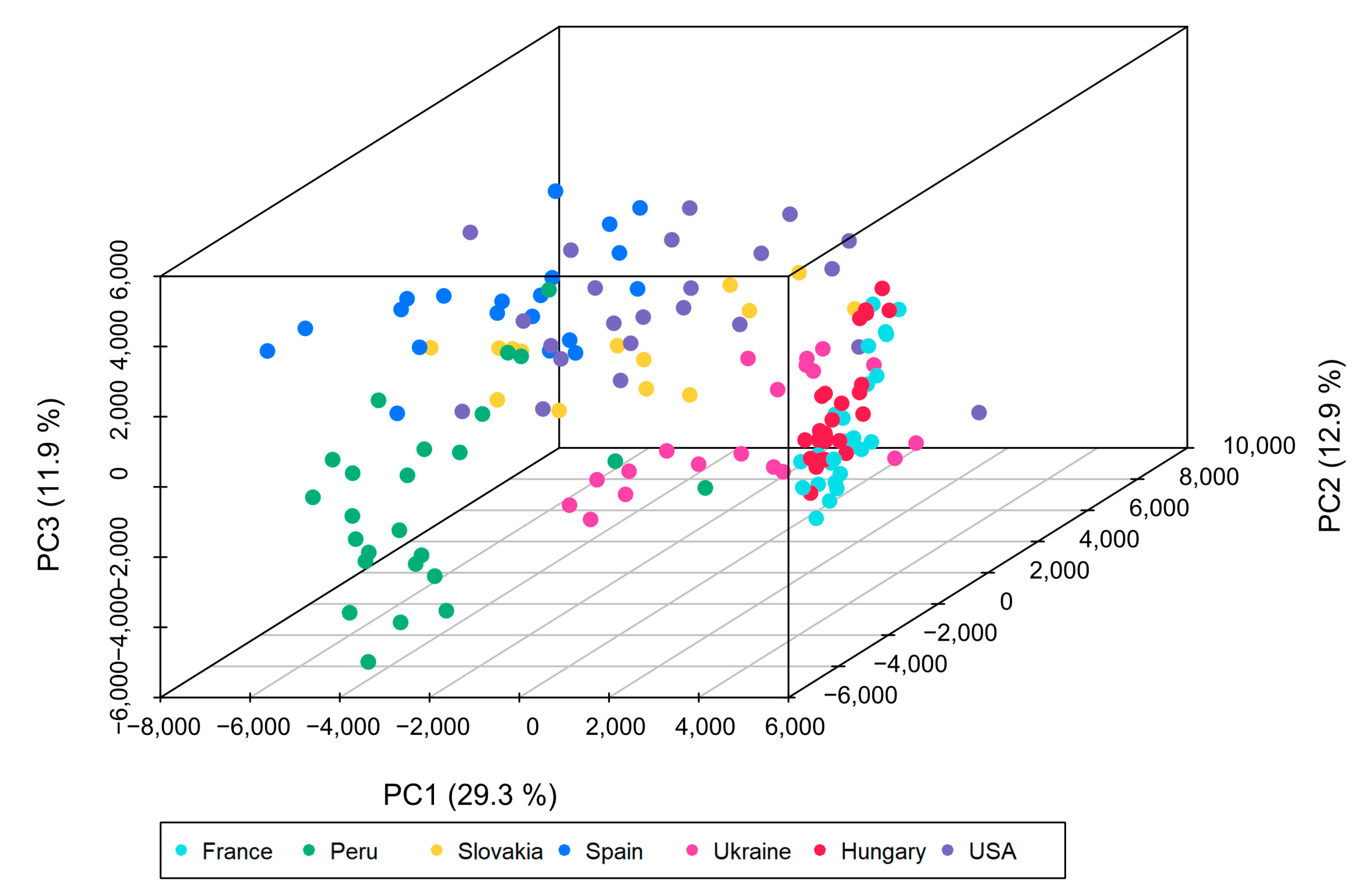
3.2. Multivariate Data Analysis
3.3. Identification of Relevant Marker Substances
- Show significance in one-way ANOVA-test regarding the geographical origin (p-value of ≤0.01);
- Have a high count of >7 out of 21 possible significant differences between country pairs determined by post-hoc Tukey test;
- Exhibit a RSD below 15%.
3.4. Structural Elucidation of Marker Metabolites
3.5. Further Investigation of the Identified Potent Marker Substances
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
References
- Creydt, M.; Fischer, M. Omics approaches for food authentication. Electrophoresis 2018, 39, 1569–1581. [Google Scholar] [CrossRef]
- Pinotti, L.; Dell’Orto, V. Feed safety in the feed supply chain. Biotechnol. Agron. Soc. Environ. 2011, 15, 9–14. [Google Scholar]
- Győri, Z. Corn: Grain-Quality Characteristics and Management of Quality Requirements. In Cereal Grains; Elsevier: Amsterdam, The Netherlands, 2017; pp. 257–290. [Google Scholar]
- Eckhoff, S.R.; Paulsen, M.R. Cereal Grain Quality; Henry, R.J., Kettlewell, P.S., Eds.; Springer: Dordrecht, The Netherlands, 1996; ISBN 978-94-010-7177-2. [Google Scholar]
- Lee, H.J.; Ryu, D. Worldwide Occurrence of Mycotoxins in Cereals and Cereal-Derived Food Products: Public Health Perspectives of Their Co-occurrence. J. Agric. Food Chem. 2017, 65, 7034–7051. [Google Scholar] [CrossRef] [PubMed]
- Esslinger, S.; Riedl, J.; Fauhl-Hassek, C. Potential and limitations of non-targeted fingerprinting for authentication of food in official control. Food Res. Int. 2014, 60, 189–204. [Google Scholar] [CrossRef]
- Scalbert, A.; Brennan, L.; Fiehn, O.; Hankemeier, T.; Kristal, B.S.; van Ommen, B.; Pujos-Guillot, E.; Verheij, E.; Wishart, D.; Wopereis, S. Mass-spectrometry-based metabolomics: Limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics 2009, 5, 435–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Ernst, M.; Silva, D.B.; Silva, R.R.; Vêncio, R.Z.N.; Lopes, N.P. Mass spectrometry in plant metabolomics strategies: From analytical platforms to data acquisition and processing. Nat. Prod. Rep. 2014, 31, 784–806. [Google Scholar] [CrossRef]
- Arena, E.; Campisi, S.; Fallico, B.; Maccarone, E. Distribution of fatty acids and phytosterols as a criterion to discriminate geographic origin of pistachio seeds. Food Chem. 2007, 104, 403–408. [Google Scholar] [CrossRef]
- Tres, A.; Ruiz-Samblas, C.; van der Veer, G.; van Ruth, S.M. Geographical provenance of palm oil by fatty acid and volatile compound fingerprinting techniques. Food Chem. 2013, 137, 142–150. [Google Scholar] [CrossRef]
- Tres, A.; Heenan, S.P.; van Ruth, S. Authentication of dried distilled grain with solubles (DDGS) by fatty acid and volatile profiling. LWT—Food Sci. Technol. 2014, 59, 215–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Creydt, M.; Hudzik, D.; Rurik, M.; Kohlbacher, O.; Fischer, M. Food Authentication: Small-Molecule Profiling as a Tool for the Geographic Discrimination of German White Asparagus. J. Agric. Food Chem. 2018, 66, 13328–13339. [Google Scholar] [CrossRef]
- Klockmann, S.; Reiner, E.; Bachmann, R.; Hackl, T.; Fischer, M. Food Fingerprinting: Metabolomic Approaches for Geographical Origin Discrimination of Hazelnuts (Corylus avellana) by UPLC-QTOF-MS. J. Agric. Food Chem. 2016, 64, 9253–9262. [Google Scholar] [CrossRef]
- Rubert, J.; Hurkova, K.; Stranska, M.; Hajslova, J. Untargeted metabolomics reveals links between Tiger nut (Cyperus esculentus L.) and its geographical origin by metabolome changes associated with membrane lipids. Food Addit. Contam. Part A 2018, 35, 1861–1869. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Shang, K.; Li, X.; Zhang, C.-H.; Liu, J.-Q.; Huang, D.-Q. Characterization and discrimination of selected China’s domestic pork using an LC-MS-based lipidomics approach. Food Control 2019, 100, 305–314. [Google Scholar] [CrossRef]
- Lim, D.K.; Mo, C.; Lee, J.H.; Long, N.P.; Dong, Z.; Li, J.; Lim, J.; Kwon, S.W. The integration of multi-platform MS-based metabolomics and multivariate analysis for the geographical origin discrimination of Oryza sativa L. J. Food Drug Anal. 2018, 26, 769–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, L.C.; Liong, C.Y.; Jemain, A.A. Partial least squares-discriminant analysis (PLS-DA) for classification of high-dimensional (HD) data: A review of contemporary practice strategies and knowledge gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef]
- Trainor, P.J.; de Filippis, A.P.; Rai, S.N. Evaluation of classifier performance for multiclass phenotype discrimination in untargeted metabolomics. Metabolites 2017, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Shiferaw, B.; Prasanna, B.M.; Hellin, J.; Bänziger, M. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 2011, 3, 307–327. [Google Scholar] [CrossRef] [Green Version]
- USDA Grain: World Markets and Trade. Available online: https://downloads.usda.library.cornell.edu/usda-esmis/files/zs25x844t/6m312f84z/3r075m960/grain.pdf (accessed on 27 May 2021).
- Montero-Vargas, J.M.; Ortíz-Islas, S.; Ramírez-Sánchez, O.; García-Lara, S.; Winkler, R. Prediction of the antioxidant capacity of maize (Zea mays) hybrids using mass fingerprinting and data mining. Food Biosci. 2020, 37, 100647. [Google Scholar] [CrossRef]
- Pastor, K.; Ačanski, M.; Vujić, Đ.; Jovanović, Đ.; Wienkoop, S. Authentication of Cereal Flours by Multivariate Analysis of GC–MS Data. Chromatographia 2016, 79, 1387–1393. [Google Scholar] [CrossRef]
- Baniasadi, H.; Vlahakis, C.; Hazebroek, J.; Zhong, C.; Asiago, V. Effect of environment and genotype on commercial maize hybrids using LC/MS-based metabolomics. J. Agric. Food Chem. 2014, 62, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Hazebroek, J.; Zhong, C.; Harp, T.; Vlahakis, C.; Baumhover, B.; Asiago, V. Effect of Genetics, Environment, and Phenotype on the Metabolome of Maize Hybrids Using GC/MS and LC/MS. J. Agric. Food Chem. 2017, 65, 5215–5225. [Google Scholar] [CrossRef] [PubMed]
- Achten, E.; Schütz, D.; Fischer, M.; Fauhl-Hassek, C.; Riedl, J.; Horn, B. Classification of Grain Maize (Zea mays L.) from Different Geographical Origins with FTIR Spectroscopy—a Suitable Analytical Tool for Feed Authentication? Food Anal. Methods 2019, 12, 2172–2184. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The human metabolome database. Nucleic Acids Res. 2007, 35, 521–526. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, 527–532. [Google Scholar] [CrossRef] [Green Version]
- Han, X. Fragmentation Patterns of Glycerolipids. In Lipidomics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 217–228. [Google Scholar]
- Pi, J.; Wu, X.; Feng, Y. Fragmentation patterns of five types of phospholipids by ultra-high-performance liquid chromatography electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Anal. Methods 2016, 8, 1319–1332. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Martin, M.M. Technical Aspects and Pitfalls of LC/MS Hyphenation. In The HPLC-MS Handbook for Practitioners; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 19–71. ISBN 9783527809202. [Google Scholar]
- De Mendiburu, F.; Yassen, M. Agricolae: Statistical Procedures for Agricultural Research. Available online: https://cran.r-project.org/package=agricolae (accessed on 8 August 2021).
- McFerrin, L. HDMD: Statistical Analysis Tools for High Dimension Molecular Data (HDMD). Available online: https://cran.r-project.org/package=HDMD (accessed on 8 August 2021).
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002; ISBN 0-387-95457-0. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Kuhn, M. Caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar]
- Kalo, P.J.; Ollilainen, V.; Rocha, J.M.; Malcata, F.X. Identification of molecular species of simple lipids by normal phase liquid chromatography—Positive electrospray tandem mass spectrometry, and application of developed methods in comprehensive analysis of low erucic acid rapeseed oil lipids. Int. J. Mass Spectrom. 2006, 254, 106–121. [Google Scholar] [CrossRef]
- Shao, B.; Li, H.; Shen, J.; Wu, Y. Nontargeted Detection Methods for Food Safety and Integrity. Annu. Rev. Food Sci. Technol. 2019, 10, 429–455. [Google Scholar] [CrossRef]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upchurch, R.G. Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett. 2008, 30, 967–977. [Google Scholar] [CrossRef]
- Murphy, D.J. Plant Storage Lipids. eLS 2016, 1–7. [Google Scholar] [CrossRef]
- Gu, Y.; He, L.; Zhao, C.; Wang, F.; Yan, B.; Gao, Y.; Li, Z.; Yang, K.; Xu, J. Biochemical and transcriptional regulation of membrane lipid metabolism in maize leaves under low temperature. Front. Plant Sci. 2017, 8, 2053. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Guo, Y.; Yan, J.; Zhang, J.; Song, T.; Rocheford, T.; Li, J.S. Major and minor QTL and epistasis contribute to fatty acid compositions and oil concentration in high-oil maize. Theor. Appl. Genet. 2010, 120, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Laurie, C.C.; Chasalow, S.D.; LeDeaux, J.R.; McCarroll, R.; Bush, D.; Hauge, B.; Lai, C.; Clark, D.; Rocheford, T.R.; Dudley, J.W. The genetic architecture of response to long-term artificial selection for oil concentration in the maize kernel. Genetics 2004, 168, 2141–2155. [Google Scholar] [CrossRef] [Green Version]
- De Abreu e Lima, F.; Li, K.; Wen, W.; Yan, J.; Nikoloski, Z.; Willmitzer, L.; Brotman, Y. Unraveling lipid metabolism in maize with time-resolved multi-omics data. Plant J. 2018, 93, 1102–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Řezanka, T.; Sigler, K. Odd-numbered very-long-chain fatty acids from the microbial, animal and plant kingdoms. Prog. Lipid Res. 2009, 48, 206–238. [Google Scholar] [CrossRef]
- Holčapek, M.; Jandera, P.; Zderadička, P.; Hrubá, L. Characterization of triacylglycerol and diacylglycerol composition of plant oils using high-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. J. Chromatogr. A 2003, 1010, 195–215. [Google Scholar] [CrossRef]



| Model | Accuracy 1 |
|---|---|
| France vs. rest | 98.9 ± 0.6% |
| Peru vs. rest | 97.0 ± 0.4% |
| Slovakia vs. rest | 91.5 ± 0.6% |
| Spain vs. rest | 96.1 ± 0.6% |
| Ukraine vs. rest | 98.5 ± 0.3% |
| Hungary vs. rest | 99.2 ± 0.3% |
| USA vs. rest | 89.3 ± 1.0% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schütz, D.; Achten, E.; Creydt, M.; Riedl, J.; Fischer, M. Non-Targeted LC-MS Metabolomics Approach towards an Authentication of the Geographical Origin of Grain Maize (Zea mays L.) Samples. Foods 2021, 10, 2160. https://doi.org/10.3390/foods10092160
Schütz D, Achten E, Creydt M, Riedl J, Fischer M. Non-Targeted LC-MS Metabolomics Approach towards an Authentication of the Geographical Origin of Grain Maize (Zea mays L.) Samples. Foods. 2021; 10(9):2160. https://doi.org/10.3390/foods10092160
Chicago/Turabian StyleSchütz, David, Elisabeth Achten, Marina Creydt, Janet Riedl, and Markus Fischer. 2021. "Non-Targeted LC-MS Metabolomics Approach towards an Authentication of the Geographical Origin of Grain Maize (Zea mays L.) Samples" Foods 10, no. 9: 2160. https://doi.org/10.3390/foods10092160






