Albedo- and Flavedo-Specific Transcriptome Profiling Related to Penicillium digitatum Infection in Citrus Fruit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal and Fruit Material
2.2. Fruit Inoculation and Disease Severity Determination
2.3. Total RNA Extraction
2.4. RNA-Seq, Data Processing and Normalization
2.5. Gene Expression Analysis
2.6. Statistical Analysis
3. Results
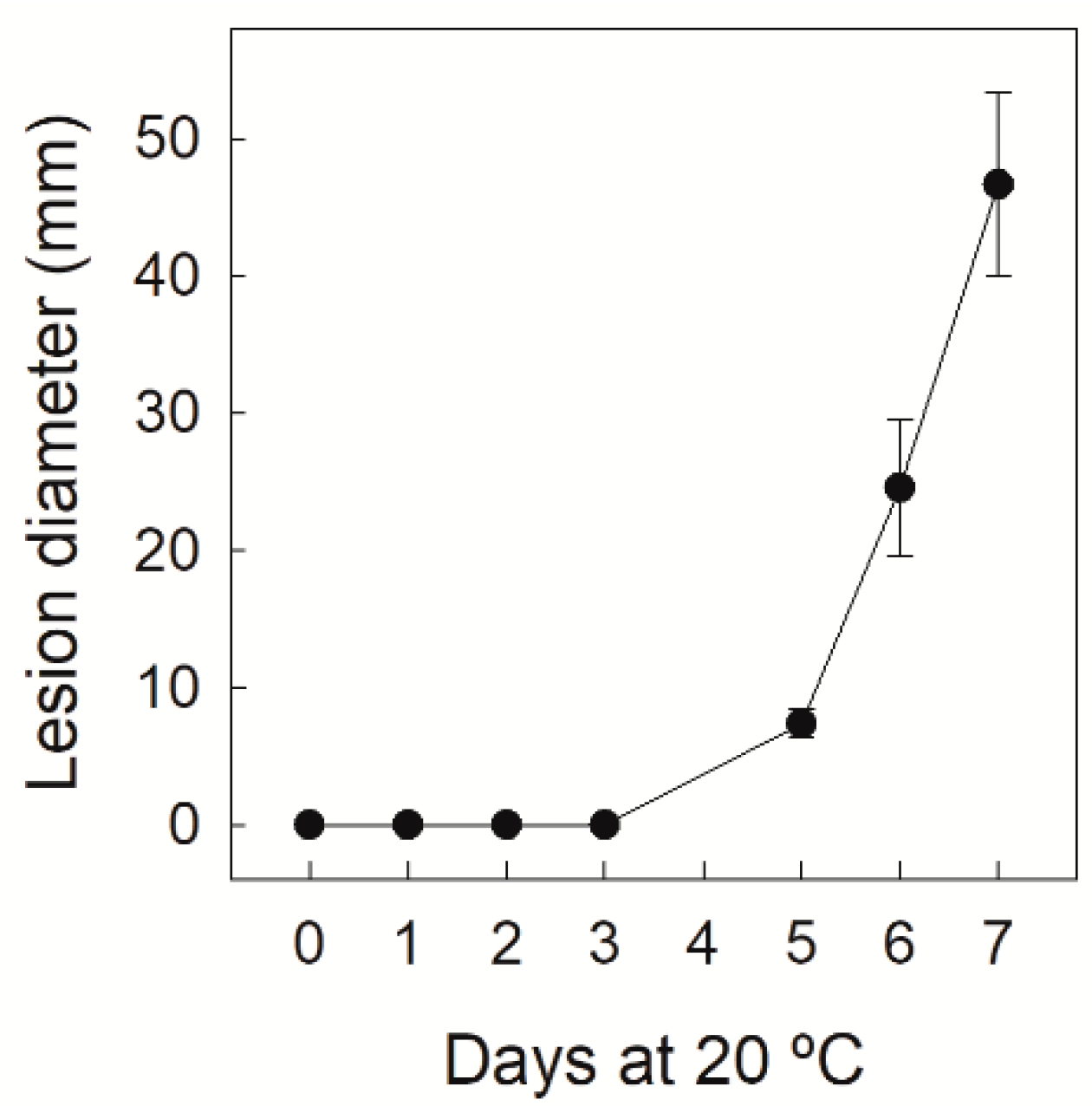
3.1. Disease Severity Evolution in Navelate Fruit Infected by P. digitatum
3.2. Comparative Transcriptional Profiling during Infection by P. digitatum between Albedo and Flavedo
3.3. Comparison of the Induced or Repressed Metabolic Pathways in Albedo in Relation to Flavedo in Navelate Orange at Fruit Harvest
3.4. Albedo and Flavedo Early Responses to P. digitatum Infection in Citrus Fruit
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alkan, N.; Fortes, A.M. Insights into molecular and metabolic events associated with fruit response to post-harvest fungal pathogens. Front. Plant Sci. 2015, 6, 889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, S.; Torres, R.; Ballester, A.-R.; Li, B.; Vilanova, L.; González-Candelas, L. Molecular aspects in pathogen-fruit interactions: Virulence and resistance. Postharvest Biol. Technol. 2016, 122, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Palou, L.; Ali, A.; Fallik, E.; Romanazzi, G. GRAS, plant- and animal-derived compounds as alternatives to conventional fungicides for the control of postharvest diseases of fresh horticultural produce. Postharvest Biol. Technol. 2016, 122, 41–52. [Google Scholar] [CrossRef]
- Ozkana, R.; Smilanickb, J.L.; Karabuluta, O.A. Toxicity of ozone gas to conidia of Penicillium digitatum, Penicillium italicum, and Botrytis cinerea and control of gray mold on table grapes. Postharvest Biol. Technol. 2011, 60, 47–51. [Google Scholar] [CrossRef]
- González-Candelas, L.; Alamar, S.; Sánchez-Torres, P.; Zacarías, L.; Marcos, J.F. A transcriptomic approach highlights induction of secondary metabolism in citrus fruit in response to Penicillium digitatum infection. BMC Plant Biol. 2010, 10, 194. [Google Scholar] [CrossRef] [Green Version]
- López-Pérez, M.; Ballester, A.-R.; González-Candelas, L. Identification and functional analysis of Penicillium digitatum genes putatively involved in virulence towards citrus fruit. Mol. Plant Pathol. 2015, 16, 262–275. [Google Scholar] [CrossRef] [Green Version]
- OuYang, Q.; Tao, N.; Guoxing, J. Transcriptional profiling analysis of Penicillium digitatum, the causal agent of citrus green mold, unravels an inhibited ergosterol biosynthesis pathway in response to citral. BMC Genom. 2016, 17, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, B.; Wang, W.; Deng, L.; Yao, S.; Ming, J.; Zeng, K. Comparative RNA-seq analysis of citrus fruit in response to infection with three major postharvest fungi. Postharvest Biol. Technol. 2018, 146, 134–146. [Google Scholar] [CrossRef]
- Qian, X.; Yang, Q.; Zhang, Q.; Abdelhai, M.H.; Dhanasekaran, S.; Serwah, B.N.A.; Gu, N.; Zhang, H. Elucidation of the initial growth process and the infection mechanism of Penicillium digitatum on postharvest citrus (Citrus reticulata Blanco). Microorganisms 2019, 7, 485. [Google Scholar] [CrossRef] [Green Version]
- Ballester, A.-R.; Lafuente, M.T.; Forment, J.; Gadea, J.; de Vos, R.C.H.; Bovy, A.G.; González-Candelas, L. Transcriptomic profiling of citrus fruit peel tissues reveals fundamental effects of phenylpropanoids and ethylene on induced resistance. Mol. Plant Pathol. 2011, 12, 879–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hershkovitz, V.; Sela, N.; Taha-Salaime, L.; Liu, J.; Rafael, G.; Kessler, C.; Aly, R.; Levy, M.; Wisniewski, M.; Droby, S. De-novo assembly and characterization of the transcriptome of Metschnikowia fructicola reveals differences in gene expression following interaction with Penicillium digitatum and grapefruit peel. BMC Genom. 2013, 14, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Chen, O.; Deng, L.; Ruan, C.; Yi, L.; Zeng, K. Pichia galeiformis induces resistance in postharvest citrus by activating the phenylpropanoid biosynthesis pathway. J. Agric. Food Chem. 2021, 69, 2619–2631. [Google Scholar] [CrossRef]
- Lafuente, M.T.; Romero, P.; Ballester, A.-R. Coordinated activation of the metabolic pathways induced by LED blue light in citrus fruit. Food Chem. 2021, 341, 128050. [Google Scholar] [CrossRef]
- Ballester, A.R.; Lafuente, M.T.; González-Candelas, L. Spatial study of antioxidant enzymes, peroxidase and phenylalanine ammonia-lyase in the citrus fruit-Penicillium digitatum interaction. Postharvest Biol. Technol. 2006, 39, 115–124. [Google Scholar] [CrossRef]
- Cajuste, J.F.; García-Breijo, F.J.; Reig-Armiñana, J.; Lafuente, M.T. Ultrastructural and histochemical analysis reveals ethylene-induced responses underlying reduced peel collapse in detached citrus fruit. Microsc. Res. Tech. 2011, 74, 970–979. [Google Scholar] [CrossRef]
- Ballester, A.-R.; Teresa Lafuente, M.; González-Candelas, L. Citrus phenylpropanoids and defence against pathogens. Part II: Gene expression and metabolite accumulation in the response of fruits to Penicillium digitatum infection. Food Chem. 2013, 136, 285–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Establés-Ortiz, B.; Romero, P.; Ballester, A.-R.; González-Candelas, L.; Lafuente, M.T. Inhibiting ethylene perception with 1-methylcyclopropene triggers molecular responses aimed to cope with cell toxicity and increased respiration in citrus fruits. Plant Physiol. Biochem. 2016, 103, 154–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, P.; Alférez, F.; Lafuente, M.T. Involvement of phospholipases and sucrose in carbon starvation-induced non-chilling peel pitting in citrus fruit. Postharvest Biol. Technol. 2020, 169, 111295. [Google Scholar] [CrossRef]
- Ballester, A.-R.; Izquierdo, A.; Lafuente, M.T.; González-Candelas, L. Biochemical and molecular characterization of induced resistance against Penicillium digitatum in citrus fruit. Postharvest Biol. Technol. 2010, 56, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Marcet-Houben, M.; Ballester, A.-R.; de la Fuente, B.; Harries, E.; Marcos, J.F.; González-Candelas, L.; Gabaldón, T. Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genom. 2012, 13, 646. [Google Scholar] [CrossRef] [Green Version]
- Lafuente, M.T.; Ballester, A.-R.; González-Candelas, L. Involvement of abscisic acid in the resistance of citrus fruit to Penicillium digitatum infection. Postharvest Biol. Technol. 2019, 154, 31–40. [Google Scholar] [CrossRef]
- Lafuente, M.T.; Alférez, F.; Romero, P. Postharvest ethylene conditioning as a tool to reduce quality loss of stored mature sweet oranges. Postharvest Biol. Technol. 2014, 94, 104–111. [Google Scholar] [CrossRef]
- Romero, P.; Lafuente, M.T. Abscisic acid deficiency alters epicuticular wax metabolism and morphology that leads to increased cuticle permeability during sweet orange (Citrus sinensis) Fruit Ripening. Front. Plant Sci. 2020, 11, 1914. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef] [Green Version]
- Weiste, C.; Dröge-Laser, W. The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 2014, 5, 3883. [Google Scholar] [CrossRef] [Green Version]
- Majda, M.; Robert, S. The role of auxin in cell wall expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef] [Green Version]
- Cajuste, J.F.; González-Candelas, L.; Veyrat, A.; García-Breijo, F.J.; Reig-Armiñana, J.; Lafuente, M.T. Epicuticular wax content and morphology as related to ethylene and storage performance of “Navelate” orange fruit. Postharvest Biol. Technol. 2010, 55, 29–35. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Wang, J.; Zhu, R.; Lu, H.; Zheng, X.; Yu, T. Transcript profiling analysis of Rhodosporidium paludigenum-mediated signalling pathways and defense responses in mandarin orange. Food Chem. 2015, 172, 603–612. [Google Scholar] [CrossRef]
- Youssef, K.; Sanzani, S.M.; Ligorio, A.; Ippolito, A.; Terry, L.A. Sodium carbonate and bicarbonate treatments induce resistance to postharvest green mould on citrus fruit. Postharvest Biol. Technol. 2014, 87, 61–69. [Google Scholar] [CrossRef]
- Romero, P.; Alférez, F.; Establés-Ortiz, B.; Lafuente, M.T. Insights into the regulation of molecular mechanisms involved in energy shortage in detached citrus fruit. Sci. Rep. 2020, 10, 1109. [Google Scholar] [CrossRef]
- Droby, S.; Eick, A.; Macarisin, D.; Cohen, L.; Rafael, G.; Stange, R.; McColum, G.; Dudai, N.; Nasser, A.; Wisniewski, M.; et al. Role of citrus volatiles in host recognition, germination and growth of Penicillium digitatum and Penicillium italicum. Postharvest Biol. Technol. 2008, 49, 386–396. [Google Scholar] [CrossRef]
- Singh, B.; Sharma, R.A. Plant terpenes: Defense responses, phylogenetic analysis, regulation and clinical applications. 3 Biotech 2015, 5, 129–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Droby, S.; Porat, R.; Cohen, L.; Weiss, B.; Shapiro, B.; Philosoph-Hadas, S.; Meir, S. Suppressing green mold decay in grapefruit with postharvest jasmonate application. J. Am. Soc. Hortic. Sci. 1999, 124, 184–188. [Google Scholar] [CrossRef] [Green Version]
- Baetsen-Young, A.; Chen, H.; Shiu, S.H.; Day, B. Contrasting transcriptional responses to Fusarium virguliforme colonization in symptomatic and asymptomatic hosts. Plant Cell 2021, 33, 224–247. [Google Scholar] [CrossRef]
- Wang, W.; Li, T.; Chen, Q.; Deng, B.; Deng, L.; Zeng, K. Transcription factor CsWRKY65 participates in the establishment of disease resistance of citrus fruits to Penicillium digitatum. J. Agric. Food Chem. 2021, 69, 5671–5682. [Google Scholar] [CrossRef]
- Balakireva, A.V.; Zamyatnin, A.A. Indispensable role of proteases in plant innate immunity. Int. J. Mol. Sci. 2018, 19, 629. [Google Scholar] [CrossRef] [Green Version]
- El Hadrami, A.; Islam, M.R.; Adam, L.R.; Daayf, F. A cupin domain-containing protein with a quercetinase activity (VdQase) regulates Verticillium dahliae’s pathogenicity and contributes to counteracting host defenses. Front. Plant Sci. 2015, 6, 440. [Google Scholar] [CrossRef] [Green Version]
- Sasidharan, R.; Voesenek, L.A.; Pierik, R. Cell wall modifying proteins mediate plant acclimatization to biotic and abiotic stresses. CRC Crit. Rev. Plant Sci. 2011, 30, 548–562. [Google Scholar] [CrossRef]
- Vilanova, L.; Torres, R.; Viñas, I.; González-Candelas, L.; Usall, J.; Fiori, S.; Solsona, C.; Teixidó, N. Wound response in orange as a resistance mechanism against Penicillium digitatum (pathogen) and P. expansum (non-host pathogen). Postharvest Biol. Technol. 2013, 78, 113–122. [Google Scholar] [CrossRef]
- Vilanova, L.; Viñas, I.; Torres, R.; Usall, J.; Jauset, A.M.; Teixidó, N. Infection capacities in the orange-pathogen relationship: Compatible (Penicillium digitatum) and incompatible (Penicillium expansum) interactions. Food Microbiol. 2012, 29, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Meraj, T.A.; Fu, J.; Raza, M.A.; Zhu, C.; Shen, Q.; Xu, D.; Wang, Q. Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes 2020, 11, 346. [Google Scholar] [CrossRef] [Green Version]
- Macarisin, D.; Cohen, L.; Eick, A.; Rafael, G.; Belausov, E.; Wisniewski, M.; Droby, S. Postharvest pathology and mycotoxins Penicillium digitatum suppresses production of hydrogen peroxide in host tissue during infection of citrus fruit. Phytopathology 2007, 97, 1491–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buron-Moles, G.; Torres, R.; Teixidó, N.; Usall, J.; Vilanova, L.; Viñas, I. Characterisation of H2O2 production to study compatible and non-host pathogen interactions in orange and apple fruit at different maturity stages. Postharvest Biol. Technol. 2015, 99, 27–36. [Google Scholar] [CrossRef]
- Klenell, M.; Morita, S.; Tiemblo-Olmo, M.; Mühlenbock, P.; Karpinski, S.; Karpinska, B. Involvement of the chloroplast signal recognition particle cpSRP43 in acclimation to conditions promoting photooxidative stress in Arabidopsis. Plant Cell Physiol. 2005, 46, 118–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daou, M.; Faulds, C.B. Glyoxal oxidases: Their nature and properties. World J. Microbiol. Biotechnol. 2017, 33, 87. [Google Scholar] [CrossRef]
- Ravichandran, S.; Stone, S.L.; Benkel, B.; Prithiviraj, B. Purple Acid Phosphatase5 is required for maintaining basal resistance against Pseudomonas syringae in Arabidopsis. BMC Plant Biol. 2013, 13, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueiredo, J.; Silva, M.S.; Figueiredo, A. Subtilisin-like proteases in plant defence: The past, the present and beyond. Mol. Plant Pathol. 2018, 19, 1017–1028. [Google Scholar] [CrossRef]
- Su, H.G.; Zhang, X.H.; Wang, T.T.; Wei, W.L.; Wang, Y.X.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Xu, Z.S.; et al. Genome-wide identification, evolution, and expression of GDSL-type esterase/lipase gene family in soybeag. Front. Plant Sci. 2020, 11, 726. [Google Scholar] [CrossRef] [PubMed]
- Claverie, J.; Balacey, S.; Lemaître-Guillier, C.; Brulé, D.; Chiltz, A.; Granet, L.; Noirot, E.; Daire, X.; Darblade, B.; Héloir, M.-C.; et al. The cell wall-derived xyloglucan is a new DAMP triggering plant immunity in Vitis vinifera and Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 1725. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.; Yan, J.; Li, Y.; Jiang, H.; Sun, J.; Chen, Q.; Li, H.; Chu, J.; Yan, C.; Sun, X.; et al. Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol. 2012, 195, 872–882. [Google Scholar] [CrossRef]
- Noordermeer, M.A.; Veldink, G.A.; Vliegenthart, J.F.G. Fatty acid hydroperoxide lyase: A plant cytochrome P450 enzyme involved in wound healing and pest resistance. ChemBioChem 2001, 2, 494–504. [Google Scholar] [CrossRef]
- Smolen, G.; Bender, J. Arabidopsis cytochrome P450 cyp83B1 mutations activate the tryptophan biosynthetic pathway. Genetics 2002, 160, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Compagnon, V.; Diehl, P.; Benveniste, I.; Meyer, D.; Schaller, H.; Schreiber, L.; Franke, R.; Pinot, F. CYP86B1 is required for very long chain ω-hydroxyacid and α,ω-dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol. 2009, 150, 1831–1843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kachroo, A.; Shanklin, J.; Whittle, E.; Lapchyk, L.; Hildebrand, D.; Kachroo, P. The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol. Biol. 2006, 63, 257–271. [Google Scholar] [CrossRef]
- Ding, S.; Zhang, J.; Yang, L.; Wang, X.; Fu, F.; Wang, R.; Zhang, Q.; Shan, Y. Changes in cuticle components and morphology of ‘Satsuma’ mandarin (Citrus unshiu) during ambient storage and their potential role on Penicillium digitatum infection. Molecules 2020, 25, 412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lafuente, M.T.; Ballester, A.-R.; Holland, N.; Cerveró, J.; Romero, P. Interrelation between ABA and phospholipases D, C and A2 in early responses of citrus fruit to Penicillium digitatum infection. Postharvest Biol. Technol. 2021, 175, 111475. [Google Scholar] [CrossRef]
- Vlot, A.C. A quest for long-distance signals: The epidermis as central regulator of pipecolic acid-associated systemic acquired resistance. J. Exp. Bot. 2021, 72, 2266–2268. [Google Scholar] [CrossRef]
- Stahl, Y.; Faulkner, C. Receptor complex mediated regulation of symplastic traffic. Trends Plant Sci. 2016, 21, 450–459. [Google Scholar] [CrossRef]
- Denancé, N.; Szurek, B.; Noël, L.D. Emerging functions of nodulin-like proteins in non-nodulating plant species. Plant Cell Physiol. 2014, 55, 469–474. [Google Scholar] [CrossRef]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 2016, 7, 735. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Pathway/Brite Map | Term Name | Enrich Factor | Corrected p-Value (BH Method) | |
|---|---|---|---|---|
| Pathway | ||||
| A09100 | Metabolism | |||
| 00500 | Starch and sucrose metabolism | 2.84 | 3.52 × 10−2 | |
| A09130 | Environmental Information Processing | 2.54 | 1.04 × 10−4 | |
| UP | B 09132 | Signal transduction | 2.44 | 4.17 × 10−4 |
| 04075 | Plant hormone signal transduction | 2.53 | 1.52 × 10−3 | |
| A09180 | Brite Hierarchies | |||
| Protein families: signaling and cellular processes | ||||
| 02000 | Transporters | 1.95 | 6.21 × 10−3 | |
| Pathway | ||||
| A09100 | Metabolism | 1.97 | 1.24 × 10−13 | |
| B 09101 | Carbohydrate metabolism | 1.57 | 1.71 × 10−3 | |
| 00640 | Propanoate metabolism | 4.30 | 1.38 × 10−3 | |
| B 09102 | Energy metabolism | 2.14 | 2.31 × 10−4 | |
| 00195 | Photosynthesis | 5.87 | 1.43 × 10−7 | |
| 00196 | Photosynthesis—antenna proteins | 8.73 | 1.07 × 10−5 | |
| B 09103 | Lipid metabolism | 2.59 | 1.46 × 10−9 | |
| 00062 | Fatty acid elongation | 4.21 | 2.45 × 10−3 | |
| 00071 | Fatty acid degradation | 4.13 | 9.09 × 10−4 | |
| 00073 | Cutin, suberine and wax biosynthesis | 5.82 | 3.69 × 10−4 | |
| 00592 | alpha-Linolenic acid metabolism | 3.55 | 2.34 × 10−3 | |
| B 09105 | Amino acid metabolism | 1.55 | 2.26 × 10−2 | |
| 00280 | Valine, leucine and isoleucine degradation | 3.12 | 9.77 × 10−3 | |
| DOWN | B 09106 | Metabolism of other amino acids | 2.19 | 1.32 × 10−3 |
| 00480 | Glutathione metabolism | 2.58 | 7.50 × 10−3 | |
| B 09109 | Metabolism of terpenoids and polyketides | 3.26 | 1.25 × 10−9 | |
| 00900 | Terpenoid backbone biosynthesis | 3.50 | 1.73 × 10−3 | |
| 00909 | Sesquiterpenoid and triterpenoid bios… | 4.58 | 7.35 × 10−3 | |
| 00900 | Terpenoid backbone biosynthesis | 3.50 | 1.73 × 10−3 | |
| B 09110 | Biosynthesis of other secondary metabolites | 3.04 | 1.26 × 10−10 | |
| 00940 | Phenylpropanoid biosynthesis | 3.27 | 8.81 × 10−8 | |
| 00945 | Stilbenoid, diarylheptanoid and gingerol bios. | 7.64 | 5.69 × 10−4 | |
| 00941 | Flavonoid biosynthesis | 6.68 | 7.83 × 10−8 | |
| A09180 | Brite Hierarchies | |||
| B 09181 | Protein families: metabolism | 1.47 | 1.65 × 10−4 | |
| 00194 | Photosynthesis proteins | 6.51 | 4.40 × 10−12 | |
| 00199 | Cytochrome P450 | 5.73 | 1.08 × 10−8 | |
| 01006 | Prenyltransferases | 3.67 | 7.00 × 10−4 | |
| 01004 | Lpid biosynthesis proteins | 2.96 | 2.29 × 10−3 | |
| Pathway | ||||
| A09100 | Metabolism | |||
| 00904 | Diterpenoid biosynthesis | 5.52 up | 3.27 × 10−2 | |
| UP/ | 4.28 down | 4.95 × 10−3 | ||
| DOWN | A09180 | Brite Hierarchies | ||
| Protein families: genetic information processing | ||||
| 03000 | Transcription factors | 2.25 up | 4.87 × 10−3 | |
| 1.82 down | 2.34 × 10−3 |
| GO Category | GO ID | Term | Up | Down | ||
|---|---|---|---|---|---|---|
| Pattern 1: Regulated by P. digitatum in the flavedo | ||||||
| BP | GO:0045038 | protein import into chloroplast thylakoi... | ↑ | 1.30 × 10−3 | ||
| BP | GO:0009416 | response to light stimulus | ↑ | 1.47 × 10−2 | ||
| BP | GO:0030244 | cellulose biosynthetic process | ↑ | 3.45 × 10−2 | ||
| MF | GO:0016760 | cellulose synthase (UDP-forming) activit... | ↑ | 2.21 × 10−2 | ||
| MF | GO:0004185 | serine-type carboxypeptidase activity | ↑ | 4.27 × 10−2 | ||
| CC | GO:0080085 | signal recognition particle, chloroplast... | ↑ | 6.70 × 10−4 | ||
| CC | GO:0009507 | chloroplast | ↑ | 4.69 × 10−3 | ||
| BP | GO:0006073 | cellular glucan metabolic process | ↓ | 2.10 × 10−3 | ||
| BP | GO:0009733 | response to auxin | ↓ | 2.60 × 10−3 | ||
| BP | GO:0009247 | glycolipid biosynthetic process | ↓ | 2.51 × 10−2 | ||
| MF | GO:0016762 | xyloglucan:xyloglucosyl transferase acti... | ↓ | 5.30 × 10−4 | ||
| MF | GO:0016787 | hydrolase activity | ↓ | 1.17 × 10−3 | ||
| CC | GO:0048046 | apoplast | ↓ | 2.40 × 10−4 | ||
| CC | GO:0005618 | cell wall | ↓ | 2.31 × 10−3 | ||
| Pattern 2: Regulated by P. digitatum in the albedo | ||||||
| BP | GO:0055114 | oxidation-reduction process | ↑ | 9.20 × 10−5 | ↓ | 2.40 × 10−3 |
| BP | GO:0006631 | fatty acid metabolic process | ↑ | 4.00 × 10−2 | ||
| MF | GO:0016705 | oxidoreductase activity, acting on paire... | ↑ | 4.50 × 10−7 | ↓ | 1.61 × 10−2 |
| MF | GO:0005506 | iron ion binding | ↑ | 1.10 × 10−6 | ↓ | 1.71 × 10−2 |
| MF | GO:0020037 | heme binding | ↑ | 4.60 × 10−6 | ↓ | 3.40 × 10−3 |
| MF | GO:0045300 | acyl-[acyl-carrier-protein] desaturase a... | ↑ | 5.40 × 10−4 | ||
| MF | GO:0046983 | protein dimerization activity | ↑ | 9.90 × 10−3 | ||
| MF | GO:0010333 | terpene synthase activity | ↑ | 4.53 × 10−2 | ||
| CC | None | |||||
| BP | GO:0000160 | phosphorelay signal transduction system | ↓ | 3.10 × 10−3 | ||
| BP | GO:0006694 | steroid biosynthetic process | ↓ | 4.85 × 10−2 | ||
| MF | GO:0005215 | transporter activity | ↓ | 2.20 × 10−3 | ||
| MF | GO:0050660 | flavin adenine dinucleotide binding | ↓ | 8.90 × 10−3 | ||
| MF | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogen... | ↓ | 3.84 × 10−2 | ||
| CC | None | |||||
| Pattern 3: Commonly regulated by P. digitatum in the albedo and flavedo | ||||||
| BP | GO:0006662 | glycerol ether metabolic process | ↓A | 2.72 × 10−2 | ||
| BP | GO:0006662 | glycerol ether metabolic process | ↓F | 4.50 × 10−2 | ||
| MF | GO:0008171 | O-methyltransferase activity | ↑A | 3.50 × 10−3 | ||
| MF | GO:0008171 | O-methyltransferase activity | ↑F | 8.05 × 10−3 | ||
| MF | GO:0045735 | nutrient reservoir activity | ↓A | 1.28 × 10−2 | ||
| MF | GO:0045735 | nutrient reservoir activity | ↓F | 1.62 × 10−2 | ||
| CC | None | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lafuente, M.T.; Romero, P.; González-Candelas, L. Albedo- and Flavedo-Specific Transcriptome Profiling Related to Penicillium digitatum Infection in Citrus Fruit. Foods 2021, 10, 2196. https://doi.org/10.3390/foods10092196
Lafuente MT, Romero P, González-Candelas L. Albedo- and Flavedo-Specific Transcriptome Profiling Related to Penicillium digitatum Infection in Citrus Fruit. Foods. 2021; 10(9):2196. https://doi.org/10.3390/foods10092196
Chicago/Turabian StyleLafuente, María Teresa, Paco Romero, and Luis González-Candelas. 2021. "Albedo- and Flavedo-Specific Transcriptome Profiling Related to Penicillium digitatum Infection in Citrus Fruit" Foods 10, no. 9: 2196. https://doi.org/10.3390/foods10092196
APA StyleLafuente, M. T., Romero, P., & González-Candelas, L. (2021). Albedo- and Flavedo-Specific Transcriptome Profiling Related to Penicillium digitatum Infection in Citrus Fruit. Foods, 10(9), 2196. https://doi.org/10.3390/foods10092196







