Prevalence, Virulence Genes, Phylogenetic Analysis, and Antimicrobial Resistance Profile of Helicobacter Species in Chicken Meat and Their Associated Environment at Retail Shops in Egypt
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. Isolation and Identification of Helicobacter spp.
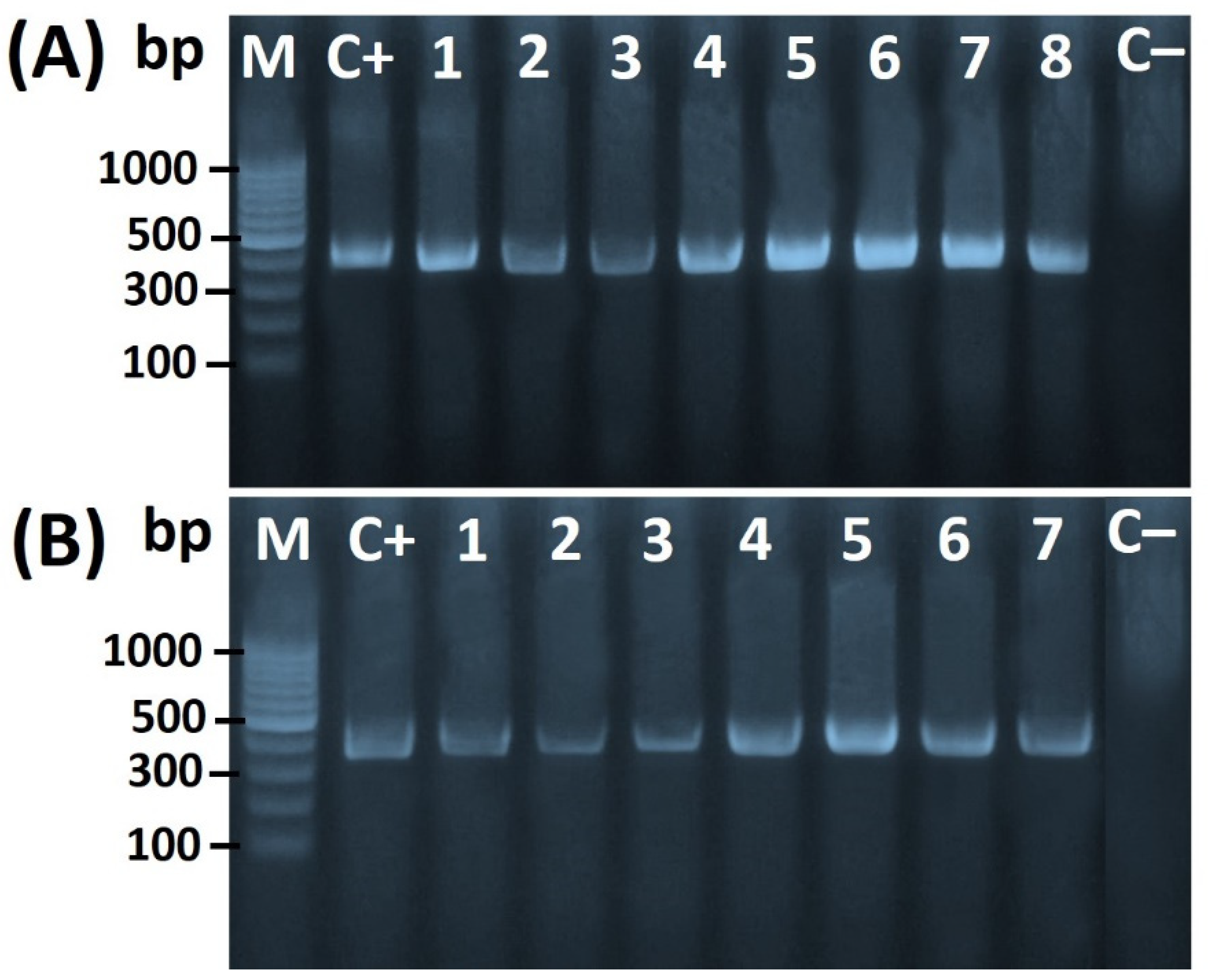
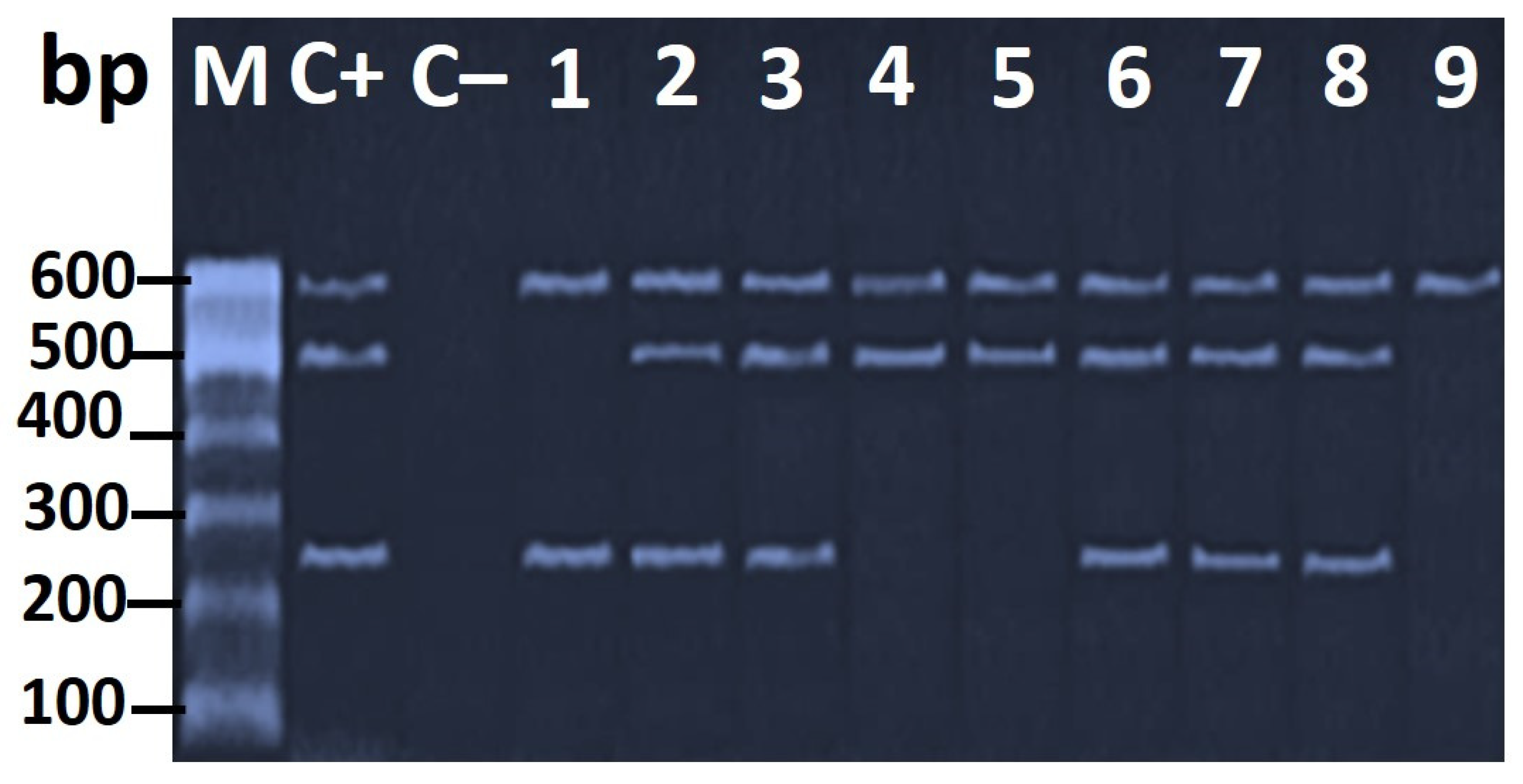
2.3. Molecular Confirmation of Helicobacter spp.
2.4. Antimicrobial Susceptibility Testing
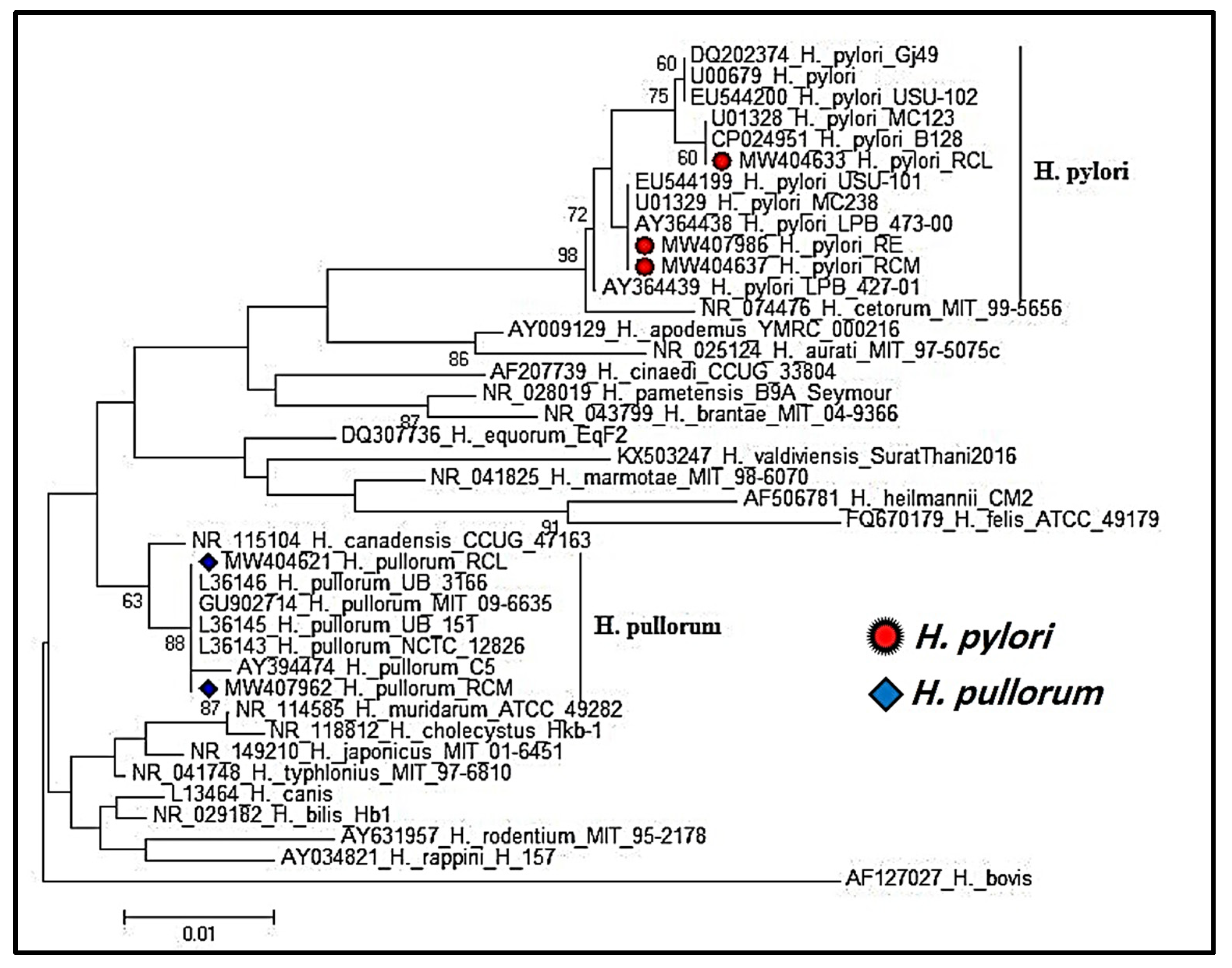
2.5. Helicobacter Species 16S rRNA Gene Sequencing and Phylogenetic Analysis
2.6. Statistical Analysis
3. Results and Discussions
3.1. Prevalence of Helicobacter spp. in Chicken Meats and Swab Samples
3.2. Phylogenetic Analysis of Partial 16S rRNA Gene Sequencing of Helicobacter Species
3.3. Genotypic Characterization of H. pylori Virulence Genes
3.4. Antimicrobial Resistance Profiles of the H. pylori and H. pullorum Isolates
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marouf, S.; Khalf, M.A.; Alorabi, M.; El-Shehawi, A.M.; El-Tahan, A.M.; Abd El-Hack, M.E.; El-Saadony, M.T.; Salem, H.M. Mycoplasma gallisepticum: A devastating organism for the poultry industry in Egypt. Poult. Sci. 2021, 101, 101658. [Google Scholar] [CrossRef] [PubMed]
- Jennings, J.L.; Sait, L.C.; Perrett, C.A.; Foster, C.; Williams, L.K.; Humphrey, T.J.; Cogan, T.A. Campylobacter jejuni is associated with, but not sufficient to cause vibrionic hepatitis in chickens. Vet. Microbiol. 2011, 149, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Qumar, S.; Majid, M.; Kumar, N.; Tiwari, S.K.; Semmler, T.; Devi, S.; Baddam, R.; Hussain, A.; Shaik, S.; Ahmed, N. Genome dynamics and molecular infection epidemiology of multidrug-resistant Helicobacter pullorum isolates obtained from broiler and free-range chickens in India. Appl. Environ. Microbiol. 2016, 83, e02305–e02316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, V.; Santos, A.; Correia, C.B.; Saraiva, M.; Ménard, A.; Vieira, L.; Sampaio, D.A.; Pinheiro, M.; Gomes, J.P.; Oleastro, M. Helicobacter pullorum isolated from fresh chicken meat: Antibiotic resistance and genomic traits of an emerging foodborne pathogen. Appl. Environ. Microbiol. 2015, 81, 8155–8163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javed, K.; Gul, F.; Abbasi, R.; Zaidi, R.A.; Noreen, Z.; Bokhari, H.; Javed, S. Prevalence and role of Type six secretion system in pathogenesis of emerging zoonotic pathogen Helicobacter pullorum from retail poultry. Avian. Pathol. 2019, 48, 557–563. [Google Scholar] [CrossRef]
- Atherton, J.C. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu. Rev. Pathol. 2006, 1, 63–96. [Google Scholar] [CrossRef]
- Kennemann, L.; Didelot, X.; Aebischer, T.; Kuhn, S.; Drescher, B.; Droege, M.; Reinhardt, R.; Correa, P.; Meyer, T.F.; Josenhans, C. Helicobacter pylori genome evolution during human infection. Proc. Natl. Acad. Sci. 2011, 108, 5033–5038. [Google Scholar] [CrossRef] [Green Version]
- Zamani, M.; Ebrahimtabar, F.; Zamani, V.; Miller, W.; Alizadeh-Navaei, R.; Shokri-Shirvani, J.; Derakhshan, M. Systematic review with meta-analysis: The worldwide prevalence of Helicobacter pylori infection. Aliment Pharmacol. Ther. 2018, 47, 868–876. [Google Scholar] [CrossRef] [Green Version]
- Ruggiero, P. Helicobacter pylori and inflammation. Curr. Pharm. Des. 2010, 16, 4225–4236. [Google Scholar] [CrossRef]
- Stanley, J.; Linton, D.; Burnens, A.P.; Dewhirst, F.E.; On, S.L.; Porter, A.; Owen, R.J.; Costas, M. Helicobacter pullorum sp. nov.-genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology 1994, 140, 3441–3449. [Google Scholar] [CrossRef] [Green Version]
- Hameed, K.G.A.; Sender, G. Prevalence of Helicobacter pullorum in Egyptian hen’s eggs and in vitro susceptibility to different antimicrobial agents. Anim. Sci. Pap. Rep. 2011, 29, 257–264. [Google Scholar]
- Atabay, H.; Corry, J.; On, S.L. Diversity and prevalence of Arcobacter spp. in broiler chickens. J. Appl. Microbiol. 1998, 84, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Wai, S.S.; Abdul-Aziz, S.; Bitrus, A.A.; Zunita, Z.; Abu, J. Helicobacter pullorum in broiler chickens and the farm environment: A One Health approach. Int. J. One Health 2019, 5, 20–25. [Google Scholar] [CrossRef] [Green Version]
- González, A.; Piqueres, P.; Moreno, Y.; Cañigral, I.; Owen, R.J.; Hernández, J.; Ferrús, M.A. A novel real-time PCR assay for the detection of Helicobacter pullorum-like organisms in chicken products. Int. Microbiol. 2008, 11, 203–208. [Google Scholar] [PubMed]
- Ceelen, L.; Decostere, A.; Verschraegen, G.; Ducatelle, R.; Haesebrouck, F. Prevalence of Helicobacter pullorum among patients with gastrointestinal disease and clinically healthy persons. J. Clin. Microbiol. 2005, 43, 2984–2986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, J. The non-H pylori helicobacters: Their expanding role in gastrointestinal and systemic diseases. Gut 2002, 50, 273–283. [Google Scholar] [CrossRef]
- Taylor, L.H.; Latham, S.M.; Woolhouse, M.E. Risk factors for human disease emergence. Philos. Trans. R Soc. Lond B Biol. Sci. 2001, 356, 983–989, Series B: Biological Sciences. [Google Scholar] [CrossRef]
- Akeel, M.; Shehata, A.; Elhafey, A.; Elmakki, E.; Aboshouk, T.; Ageely, H.; Mahfouz, M. Helicobacter pylori vacA, cagA and iceA genotypes in dyspeptic patients from southwestern region, Saudi Arabia: Distribution and association with clinical outcomes and histopathological changes. BMC Gastroenterol. 2019, 19, 16. [Google Scholar] [CrossRef]
- Sedaghat, H.; Moniri, R.; Jamali, R.; Arj, A.; Zadeh, M.R.; Moosavi, S.G.A.; Rezaei, M. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2, and oipA genotypes in patients with upper gastrointestinal diseases. Iran. J. Microbiol. 2014, 6, 14–21. [Google Scholar]
- Yong, X.; Tang, B.; Li, B.-S.; Xie, R.; Hu, C.-J.; Luo, G.; Qin, Y.; Dong, H.; Yang, S.-M. Helicobacter pylori virulence factor cagA promotes tumorigenesis of gastric cancer via multiple signaling pathways. Cell Commun. Signal. 2015, 13, 30. [Google Scholar] [CrossRef] [Green Version]
- Foegeding, N.J.; Caston, R.R.; McClain, M.S.; Ohi, M.D.; Cover, T.L. An overview of Helicobacter pylori vacA toxin biology. Toxins 2016, 8, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, T.; Wassenaar, T.M.; Peek, R.M.; Aras, R.A.; Tschumi, A.I.; van Doorn, L.-J.; Kusugami, K.; Blaser, M.J. A Helicobacter pylori restriction endonuclease-replacing gene, hrgA, is associated with gastric cancer in Asian strains. Cancer Res. 2002, 62, 2385–2389. [Google Scholar]
- Goderska, K.; Pena, S.A.; Alarcon, T. Helicobacter pylori treatment: Antibiotics or probiotics. Appl. Microbiol. Biotechnol. 2018, 102, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Alba, C.; Blanco, A.; Alarcón, T. Antibiotic resistance in Helicobacter pylori. Curr. Opin. Infect. Dis. 2017, 30, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Esaki, M.; Kusano, C.; Ikehara, H.; Gotoda, T. Development of Helicobacter pylori treatment: How do we manage antimicrobial resistance? World J. Gastroenterol. 2019, 25, 1907–1912. [Google Scholar] [CrossRef]
- Yousefi-Avarvand, A.; Vaez, H.; Tafaghodi, M.; Sahebkar, A.H.; Arzanlou, M.; Khademi, F. Antibiotic resistance of Helicobacter pylori in Iranian children: A systematic review and meta-analysis. Microb. Drug Resist. 2018, 24, 980–986. [Google Scholar] [CrossRef]
- El Dairouty, R.; Murad, H.; El Shenawy, M.; Hosny, I.; Okda, A.; El Shamy, S. Helicobacter pylori and its interrelations with other foodborne pathogenic bacteria in Egyptian meat and some meat products. Curr. Sci. Int. 2016, 5, 139–146. [Google Scholar]
- Hamada, M.; Elbehiry, A.; Marzouk, E.; Moussa, I.M.; Hessain, A.M.; Alhaji, J.H.; Heme, H.A.; Zahran, R.; Abdeen, E. Helicobacter pylori in a poultry slaughterhouse: Prevalence, genotyping and antibiotic resistance pattern. Saudi J. Biol. Sci. 2018, 25, 1072–1078. [Google Scholar] [CrossRef]
- Hassan, A.K.; Shahata, M.A.; Refaie, E.M.; Ibrahim, R.S. Detection and identification of Helicobacter pullorum in poultry species in upper Egypt. J. Adv. Vet. Res. 2014, 4, 42–48. [Google Scholar]
- Tabrizi, S.A.; Derakhshandeh, A.; Esfandiari, A.; Atashi, A.Z. Identification of Helicobacter spp. in gastrointestinal tract, pancreas and hepatobiliary system of stray cats. Iran J. Vet. Res. 2015, 16, 374–376. [Google Scholar]
- Safaei, H.; Rahimi, E.; Zandi, A.; Rashidipour, A. Helicobacter pylori as a zoonotic infection: The detection of H. pylori antigens in the milk and faeces of cows. J. Res. Med. Sci. 2011, 16, 184–187. [Google Scholar] [PubMed]
- Tiwari, S.; Khan, A.; Manoj, G.; Ahmed, S.; Abid, Z.; Habeeb, A.; Abid, Z.; Habibullah, C. A simple multiplex PCR assay for diagnosing virulent Helicobacter pylori infection in human gastric biopsy specimens from subjects with gastric carcinoma and other gastro-duodenal diseases. J. Appl. Microbiol. 2007, 103, 2353–2360. [Google Scholar] [CrossRef] [PubMed]
- CLSI 2015; Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement. CLSI document M100-S25; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015.
- EUCAST 2020; The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0; The European Society of Clinical Microbiology and Infectious Diseases (ESCMID): Basel, Switzerland, 2020. Available online: http://www.eucast.org (accessed on 23 April 2020).
- CLSI 2005; Performance Standards for Antimicrobial Susceptibility Testing. Fifteenth Informational Supplement. M100-S15; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2005.
- Zanoni, R.G.; Rossi, M.; Giacomucci, D.; Sanguinetti, V.; Manfreda, G. Occurrence and antibiotic susceptibility of Helicobacter pullorum from broiler chickens and commercial laying hens in Italy. Int. J. Food Microbiol. 2007, 116, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Zhang, H.; Law, J.; Tsang, R.; Tsang, T. Detection of Helicobacter pylori from food sources by a novel multiplex PCR assay. Food Saf. 2008, 28, 609–619. [Google Scholar] [CrossRef]
- Gholami-Ahangaran, M.; Haddadi, I.; Karimi, Y.; Omrani, E. Molecular evidence of Helicobacter pullorum, as a foodborne pathogen in broiler carcasses in Iran. Europ. Poult. Sci. 2015, 79. [Google Scholar] [CrossRef]
- Jebellijavan, A.; Emadi Chashmi, S.H.; Staji, H.; Akhlaghi, H. Comparison of the Culture and PCR Methods to Determine th Prevalence and Antibiotic Resistance of Helicobacter pullorum Isolated from Chicken Thigh Samples in Semnan, Iran. J. Hum. Environ. Health Promot. 2020, 6, 167–172. [Google Scholar] [CrossRef]
- Ceelen, L.M.; Decostere, A.; Van den Bulck, K.; On, S.L.W.; Baele, M.; Ducatelle, R.; Haesebrouck, F. Helicobacter pullorum in Chickens, Belgium. Emerg. Infect. Dis. 2006, 12, 263–267. [Google Scholar] [CrossRef] [Green Version]
- Hemmatinezhad, B.; Momtaz, H.; Rahimi, E. vacA, cagA, iceA and oipA genotypes status and antimicrobial resistance properties of Helicobacter pylori isolated from various types of ready to eat foods. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Gilani, A.; Razavilar, V.; Rokni, N.; Rahimi, E. vacA and cagA genotypes status and antimicrobial resistance properties of Helicobacter pylori strains isolated from meat products in Isfahan province, Iran. Iran J. Vet. Res. 2017, 18, 97–102. [Google Scholar] [PubMed]
- Mousavi, S.; Dehkordi, F.S. Virulence factors and antibiotic resistance of Helicobacter pylori isolated from raw milk and unpasteurized dairy products in Iran. J. Venom. Anim. Toxins Incl. Trop. Dis. 2018, 20, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjbar, R.; Yadollahi Farsani, F.; Safarpoor Dehkordi, F. Antimicrobial resistance and genotyping of vacA, cagA, and iceA alleles of the Helicobacter pylori strains isolated from traditional dairy products. J. Food Saf. 2019, 39, e12594. [Google Scholar] [CrossRef]
- Baj, J.; Forma, A.; Sitarz, M.; Portincasa, P.; Garruti, G.; Krasowska, D.; Maciejewski, R. Helicobacter pylori virulence factors—mechanisms of bacterial pathogenicity in the gastric microenvironment. Cells 2021, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Idowu, A.; Mzukwa, A.; Harrison, U.; Palamides, P.; Haas, R.; Mbao, M.; Mamdoo, R.; Bolon, J.; Jolaiya, T.; Smith, S. Detection of Helicobacter pylori and its virulence genes (cag A, dup A, and vac A) among patients with gastroduodenal diseases in Chris Hani Baragwanath Academic Hospital, South Africa. BMC Gastroenterol. 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Ando, T.; Aras, R.; Kusugami, K.; Blaser, M.; Wassenaar, T. Evolutionary history of hrgA, Which Replaces the Restriction Gene hpy IIIR in the hpy III Locus of Helicobacter pylori. J. Bacteriol. 2003, 185, 295–301. [Google Scholar] [CrossRef] [Green Version]
- Mashak, Z.; Jafariaskari, S.; Alavi, I.; Shahreza, M.S.; Dehkordi, F.S. Phenotypic and genotypic assessment of antibiotic resistance and genotyping of vacA, cagA, iceA, oipA, cagE, and babA2 alleles of Helicobacter pylori bacteria isolated from raw meat. Infect Drug Resist. 2020, 13, 257–272. [Google Scholar] [CrossRef] [Green Version]
- Secka, O.; Berg, D.E.; Antonio, M.; Corrah, T.; Tapgun, M.; Walton, R.; Thomas, V.; Galano, J.J.; Sancho, J.; Adegbola, R.A. Antimicrobial susceptibility and resistance patterns among Helicobacter pylori strains from The Gambia, West Africa. Antimicrob. Agents Chemother. 2013, 57, 1231–1237. [Google Scholar] [CrossRef] [Green Version]
- Yahaghi, E.; Khamesipour, F.; Mashayekhi, F.; Safarpoor Dehkordi, F.; Sakhaei, M.H.; Masoudimanesh, M.; Khameneie, M.K. Helicobacter pylori in vegetables and salads: Genotyping and antimicrobial resistance properties. BioMed. Res. Int. 2014, 2014, 757941. [Google Scholar] [CrossRef] [Green Version]
- Hunt, R.; Xiao, S.; Megraud, F. World Gastroenterology Organization (WGO) global guideline. Helicobacter pylori in developing countries. . J. Gastrointestin. Liver Dis. 2011, 20, 299–304. [Google Scholar] [CrossRef]
- Sallam, K.I.; Abd-Elghany, S.M.; Imre, K.; Morar, A.; Herman, V.; Hussein, M.A.; Mahros, M.A. Ensuring safety and improving keeping quality of meatballs by addition of sesame oil and sesamol as natural antimicrobial and antioxidant agents. Food Microbiol. 2021, 99, 103834. [Google Scholar] [CrossRef] [PubMed]


| Target Gene | Primers Sequences (5′–3′) | Product Size (bp) | Reference |
|---|---|---|---|
| Helicobacter spp. 16S rRNA | 5′-AAGGATGAAGCTTCTAGCTTGCTA-3′ 5′-GTGCTTATTCGTGAGATACCGTCAT-3′ | 398 | Tabrizi et al. [30] |
| Helicobacter pullorum (H. pullorum)-specific 16S rRNA | 5′-ATG AAT GCTAGTTGTTGTCAG-3′ 5′-GATTGGCTCCACTTCACA-3′ | 447 | Stanley et al. [10] |
| Helicobacter pylori (H. pylori)-specific phosphoglucosamine mutase gene (glmM) | 5′-GAATAAGCTTTTAGGGGTGTTAGGGG-3′ 5′-GCTTACTTTCTAACACTAACGCGC-3′ | 294 | Safaei et al. [31] |
| Restriction endonuclease-replacing gene A (hrgA) | 5′-TCTCGTGAAAGAGAATTTCC-3′ 5′-TAAGTGTGGGTATATCAATC-3′ | 594 | Tiwari et al. [32] |
| Cytotoxin-associated gene A (cagA) | 5′-GCGATTGTTATTGTGCTTGTAG-3′ 5′-GAAGTGGTTAAAAAACAATGCCCC-3′ | 499 | |
| Vacuolating cytotoxin A (vacA) | 5′-ATGGAAATACAACAAACACAC-3′ 5′-CTGCTTGAATGCGCCAAAC-3′ | 259 |
| Source | Sample Type | Helicobacter spp. | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| H. pylori | H. pullorum | Others | |||||||
| No. | % | No. | % | No. | % | No. | % | ||
| Retail chicken (n = 300) | Breast meat (100) | 4 | 4 | 2 | 2 | 0 | 0 | 6 | 6 |
| Liver (100) | 10 | 10 | 6 | 6 | 4 | 4 | 20 | 20 | |
| Gizzard (100) | 2 | 2 | 6 | 6 | 2 | 2 | 10 | 10 | |
| Environmental swabs (n = 30) * | Cutting boards (10) | 2 | 20 | 0 | 0 | 0 | 0 | 2 | 20 |
| Knives (10) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Workers’ hands (10) | 0 | 0 | 0 | 0 | 1 | 10 | 1 | 10 | |
| Total | 330 | 18 | 5.45 | 14 | 4.24 | 7 | 2.12 | 39 | 11.82 |
| Gene | Isolate spp. | Isolate ID | Source of Isolates | Accession Number |
|---|---|---|---|---|
| 16S rRNA | H. pylori | H. pylori_RCM | Chicken meat | MW404637 |
| H. pylori_RCL | Chicken liver | MW404633 | ||
| H. pylori_RE | Retail shop environment (Cutting board swabs) | MW407986 | ||
| H. pullorum | H. pullorum_RCM | Chicken Meat | MW407962 | |
| H. pullorum_RCL | Chicken Liver | MW404621 |
| Source | Number of Isolates | vacAs1 | cagA | hrgA | |||
|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Number | % | ||
| Breast | 4 | 4 | 100 | 2 | 50 | 4 | 100 |
| Liver | 10 | 6 | 60 | 8 | 80 | 10 | 100 |
| Gizzard | 2 | 0 | 0 | 2 | 100 | 2 | 100 |
| Environment | 2 | 2 | 100 | 2 | 100 | 2 | 100 |
| Total | 18 | 12 | 66.7 | 14 | 77.8 | 18 | 100 |
| Isolates | Antibiotic | Bp | Isolates Number According to the Results of MIC (µg/mL) | ABR No. (%) | MDR No. (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ˂0.12 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | ˃256 | |||||
| H. pylori (n = 18) | Amoxicillin | ˃0.125 | 14 | 2 | 2 | 2 (11.1) | 2 (22.2) | |||||||||||
| Clarithromycin | ≥1 | 2 | 2 | 6 | 2 | 2 | 2 | 2 | 8 (44.4) | |||||||||
| Metronidazole | ˃8 | 3 | 1 | 1 | 3 | 4 | 4 | 2 | 6 (33.3) | |||||||||
| Tetracycline | >1 | 4 | 3 | 7 | 2 | 2 | 4 (22.2) | |||||||||||
| Levofloxacin | >1 | 4 | 3 | 3 | 4 | 2 | 2 | 4 (22.2) | ||||||||||
| H. pullorum (n = 14) | Ampicillin | ≥32 | 2 | 3 | 7 | 2 | 0 (0) | 3 (42.9) | ||||||||||
| Erythromycin | ≥8 | 2 | 1 | 3 | 2 | 3 | 3 | 12 (85.7) | ||||||||||
| Tetracycline | ≥16 | 3 | 5 | 3 | 1 | 2 | 6 (42.9) | |||||||||||
| Ciprofloxacin | ≥4 | 1 | 3 | 3 | 1 | 2 | 4 | 10 (71.4) | ||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elrais, A.M.; Arab, W.S.; Sallam, K.I.; Elmegid, W.A.; Elgendy, F.; Elmonir, W.; Imre, K.; Morar, A.; Herman, V.; Elaadli, H. Prevalence, Virulence Genes, Phylogenetic Analysis, and Antimicrobial Resistance Profile of Helicobacter Species in Chicken Meat and Their Associated Environment at Retail Shops in Egypt. Foods 2022, 11, 1890. https://doi.org/10.3390/foods11131890
Elrais AM, Arab WS, Sallam KI, Elmegid WA, Elgendy F, Elmonir W, Imre K, Morar A, Herman V, Elaadli H. Prevalence, Virulence Genes, Phylogenetic Analysis, and Antimicrobial Resistance Profile of Helicobacter Species in Chicken Meat and Their Associated Environment at Retail Shops in Egypt. Foods. 2022; 11(13):1890. https://doi.org/10.3390/foods11131890
Chicago/Turabian StyleElrais, Amina Mohamed, Walid S. Arab, Khalid Ibrahim Sallam, Walaa Abd Elmegid, Fatma Elgendy, Walid Elmonir, Kálmán Imre, Adriana Morar, Viorel Herman, and Haitham Elaadli. 2022. "Prevalence, Virulence Genes, Phylogenetic Analysis, and Antimicrobial Resistance Profile of Helicobacter Species in Chicken Meat and Their Associated Environment at Retail Shops in Egypt" Foods 11, no. 13: 1890. https://doi.org/10.3390/foods11131890
APA StyleElrais, A. M., Arab, W. S., Sallam, K. I., Elmegid, W. A., Elgendy, F., Elmonir, W., Imre, K., Morar, A., Herman, V., & Elaadli, H. (2022). Prevalence, Virulence Genes, Phylogenetic Analysis, and Antimicrobial Resistance Profile of Helicobacter Species in Chicken Meat and Their Associated Environment at Retail Shops in Egypt. Foods, 11(13), 1890. https://doi.org/10.3390/foods11131890










