Lactococcus garvieae FUA009, a Novel Intestinal Bacterium Capable of Producing the Bioactive Metabolite Urolithin A from Ellagic Acid
Abstract
:1. Introduction
2. Material and Methods
2.1. Chemicals and Solvents
2.2. Isolation of UA-Producing Bacteria from Intestinal Microbiota
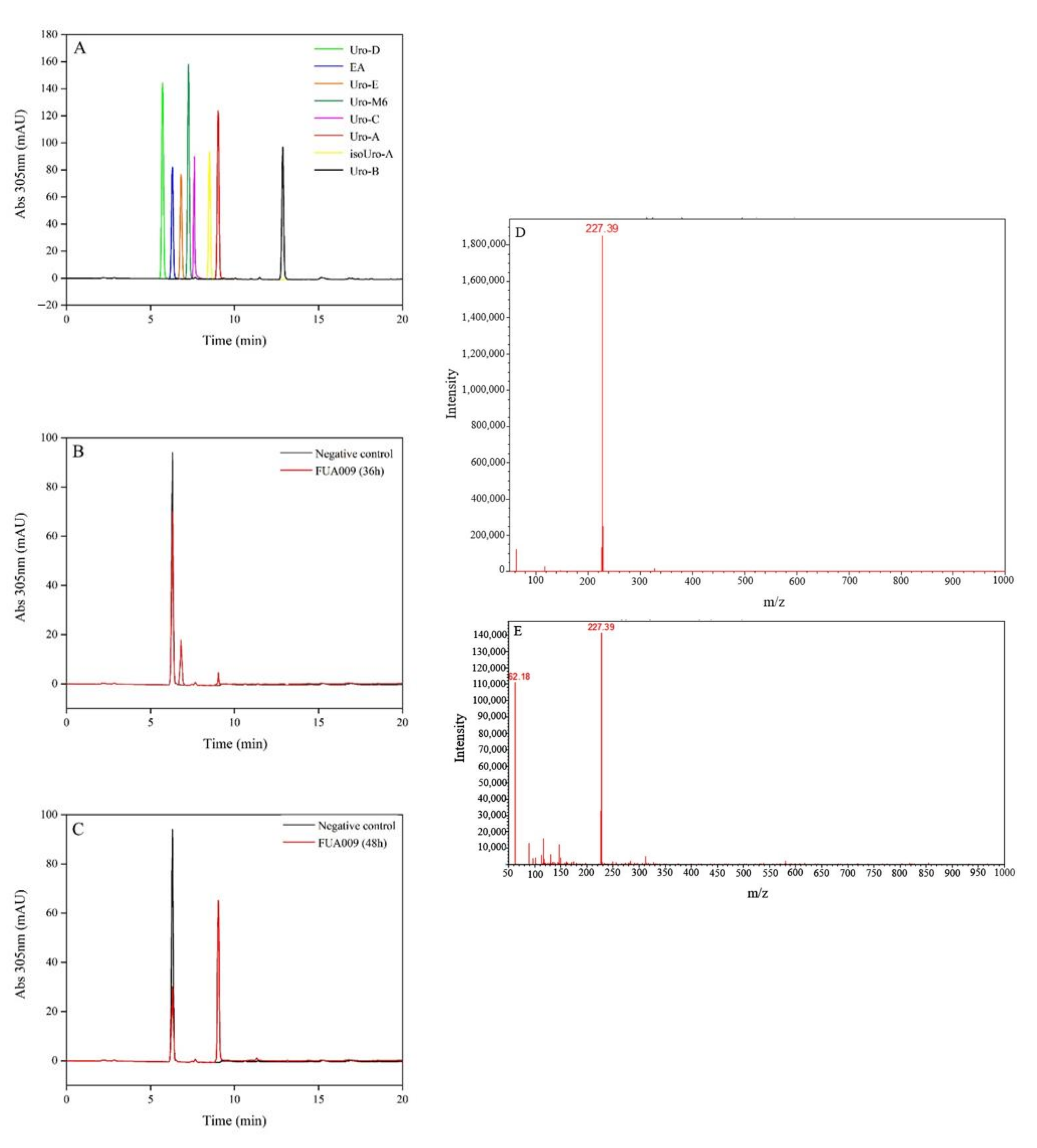
2.3. HPLC and UPLC-MS Analysis
2.4. Identification of the UA-Producing Bacteria
2.5. Whole-Genome Sequencing, Assembly, and Annotation
2.6. Safety Assessment of the UA-Producing Bacterium FUA009
2.6.1. Hemolysis Activity and Antibiotic Susceptibility Assay
2.6.2. Genome Mining for the Safety-Related Genes and Mobile Genetic Elements
2.7. Probiotic Characteristics Assessment of the UA-Producing Bacterium Lactococcus garvieae FUA009
2.7.1. Identification of Probiotic Related Genes in the Lactococcus garvieae FUA009 Genome
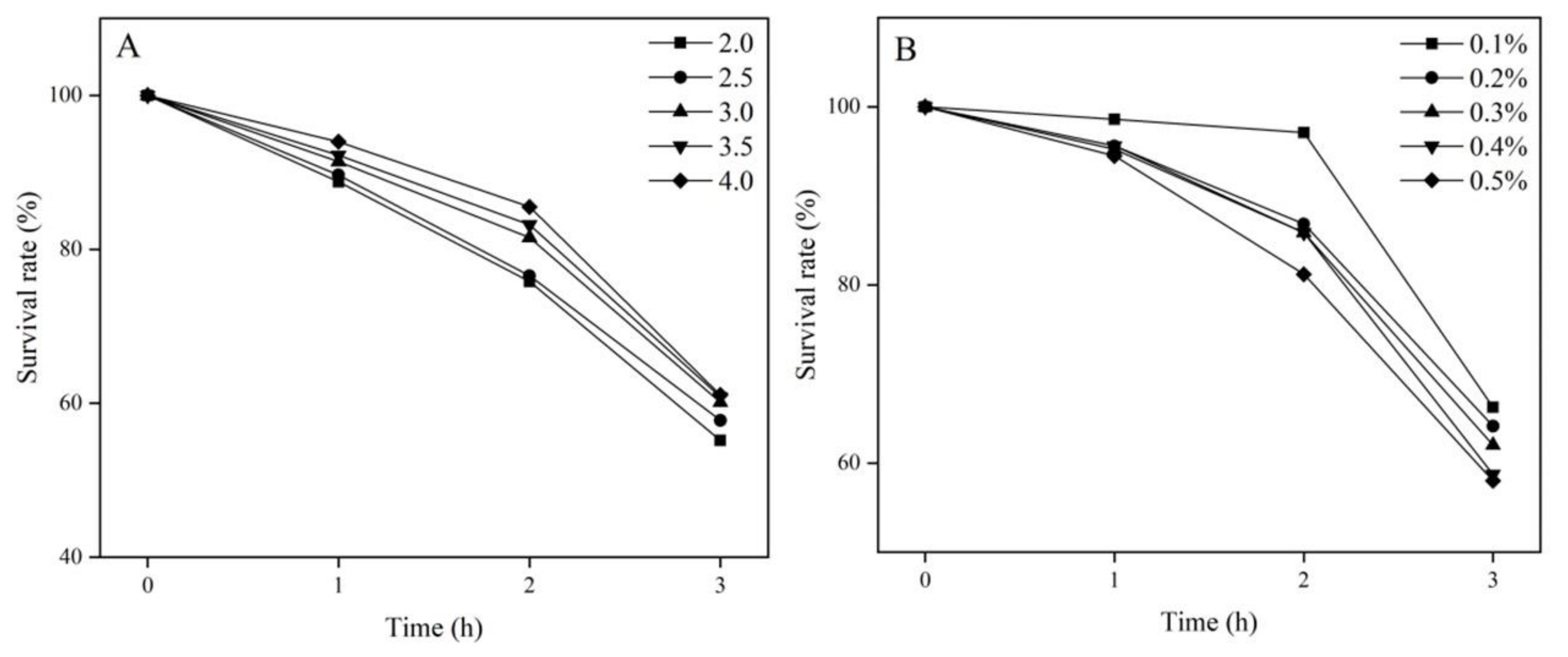
2.7.2. Evaluation of the Acid and Bile Salt Tolerance In Vitro
3. Results
3.1. Isolation and Identification of UA-Producing Bacteria
3.2. Evaluation Safety of Lactococcus garvieae FUA009
3.2.1. Antibiotic Resistance Gene Analysis in the Genome and In Vitro
3.2.2. Hemolysis Assay of Lactococcus garvieae FUA009
3.2.3. Safety-Related Gene Evaluation in the Genome of FUA009
3.3. Assessment of Probiotic Properties of Lactococcus garvieae FUA009
3.3.1. Evaluation of Stress-Responsive Protein Genes in the Lactococcus garvieae FUA009 Genome
3.3.2. Evaluation of Adhesion-Related Genes in Lactococcus garvieae FUA009 Genome
3.3.3. Tolerance of Lactococcus garvieae FUA009 to Acid and Bile In Vitro
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Doyle, B.; Griffiths, L.A. The metabolism of ellagic acid in the rat. Xenobiotica 1980, 10, 247–256. [Google Scholar] [CrossRef]
- Espin, J.C.; Larrosa, M.; Garcia-Conesa, M.T.; Tomas-Barberan, F. Biological significance of urolithins, the gut microbial ellagic acid-derived metabolites: The evidence so far. Evid. Based Complement. Alternat. Med. 2013, 2013, 270418. [Google Scholar] [CrossRef]
- Tang, L.; Mo, Y.; Li, Y.; Zhong, Y.; He, S.; Zhang, Y.; Tang, Y.; Fu, S.; Wang, X.; Chen, A. Urolithin A alleviates myocardial ischemia/reperfusion injury via PI3K/Akt pathway. Biochem. Biophys. Res. Commun. 2017, 486, 774–780. [Google Scholar] [CrossRef]
- Andreux, P.A.; Blanco-Bose, W.; Ryu, D.; Burdet, F.; Ibberson, M.; Aebischer, P.; Auwerx, J.; Singh, A.; Rinsch, C. The mitophagy activator urolithin A is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Nat. Metab. 2019, 1, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Sun, S.; Ma, G.; Hou, H.; Ma, Q.; Zhang, L.; Zhang, Z.; Wang, H.; Ying, Z. Gefitinib facilitates PINK1/Parkin-mediated mitophagy by enhancing mitochondrial recruitment of OPTN. Fundam. Res. 2022, 2022, 2667–3258. [Google Scholar] [CrossRef]
- Avila-Galvez, M.A.; Gimenez-Bastida, J.A.; Gonzalez-Sarrias, A.; Espin, J.C. Tissue deconjugation of urolithin A glucuronide to free urolithin A in systemic inflammation. Food Funct. 2019, 10, 3135–3141. [Google Scholar] [CrossRef]
- Oh, C.M.; Ryu, D.; Cho, S.; Jang, Y. Mitochondrial quality control in the heart: New drug targets for cardiovascular disease. Korean Circ. J. 2020, 50, 395–405. [Google Scholar] [CrossRef]
- Ryu, D.; Mouchiroud, L.; Andreux, P.A.; Katsyuba, E.; Moullan, N.; Nicolet-Dit-Felix, A.A.; Williams, E.G.; Jha, P.; Lo Sasso, G.; Huzard, D.; et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat. Med. 2016, 22, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, M.K.; Choudhury, S.; Borah, A. An in silico investigation on the inhibitory potential of the constituents of pomegranate juice on antioxidant defense mechanism: Relevance to neurodegenerative diseases. IBRO Rep. 2019, 6, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Ezzat-Zadeh, Z.; Henning, S.M.; Yang, J.; Woo, S.L.; Lee, R.P.; Huang, J.; Thames, G.; Gilbuena, I.; Tseng, C.H.; Heber, D.; et al. California strawberry consumption increased the abundance of gut microorganisms related to lean body weight, health and longevity in healthy subjects. Nutr. Res. 2021, 85, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cui, S.; Mao, B.; Zhang, Q.; Zhao, J.; Zhang, H.; Tang, X.; Chen, W. Ellagic acid and intestinal microflora metabolite urolithin A: A review on its sources, metabolic distribution, health benefits, and biotransformation. Crit. Rev. Food Sci. Nutr. 2022, 2022, 1–23. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Halemahebai, G.; Tian, L.; Dong, H.; Aisker, G. Urolithin A, a pomegranate metabolite, protects pancreatic beta cells from apoptosis by activating autophagy. J. Ethnopharmacol. 2021, 272, 113628. [Google Scholar] [CrossRef] [PubMed]
- Hasheminezhad, S.H.; Boozari, M.; Iranshahi, M.; Yazarlu, O.; Sahebkar, A.; Hasanpour, M.; Iranshahy, M. A mechanistic insight into the biological activities of urolithins as gut microbial metabolites of ellagitannins. Phytother. Res. 2021, 36, 112–146. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Villalba, R.; Beltran, D.; Espin, J.C.; Selma, M.V.; Tomas-Barberan, F.A. Time course production of urolithins from ellagic acid by human gut microbiota. J. Agric. Food Chem. 2013, 61, 8797–8806. [Google Scholar] [CrossRef]
- Cortes-Martin, A.; Garcia-Villalba, R.; Gonzalez-Sarrias, A.; Romo-Vaquero, M.; Loria-Kohen, V.; Ramirez-de-Molina, A.; Tomas-Barberan, F.A.; Selma, M.V.; Espin, J.C. The gut microbiota urolithin metabotypes revisited: The human metabolism of ellagic acid is mainly determined by aging. Food Funct. 2018, 9, 4100–4106. [Google Scholar] [CrossRef] [PubMed]
- Tomas-Barberan, F.A.; Garcia-Villalba, R.; Gonzalez-Sarrias, A.; Selma, M.V.; Espin, J.C. Ellagic acid metabolism by human gut microbiota: Consistent observation of three urolithin phenotypes in intervention trials, independent of food source, age, and health status. J. Agric. Food Chem. 2014, 62, 6535–6538. [Google Scholar] [CrossRef]
- Cortes-Martin, A.; Selma, M.V.; Tomas-Barberan, F.A.; Gonzalez-Sarrias, A.; Espin, J.C. Where to look into the puzzle of polyphenols and health? The postbiotics and gut microbiota associated with human metabotypes. Mol. Nutr. Food Res. 2020, 64, e1900952. [Google Scholar] [CrossRef]
- Selma, M.V.; Tomas-Barberan, F.A.; Beltran, D.; Garcia-Villalba, R.; Espin, J.C. Gordonibacter urolithinfaciens sp. nov., a urolithin-producing bacterium isolated from the human gut. Int. J. Syst. Evol. Microbiol. 2014, 64, 2346–2352. [Google Scholar] [CrossRef]
- Selma, M.V.; Beltran, D.; Garcia-Villalba, R.; Espin, J.C.; Tomas-Barberan, F.A. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Funct. 2014, 5, 1779–1784. [Google Scholar] [CrossRef]
- Selma, M.V.; Beltran, D.; Luna, M.C.; Romo-Vaquero, M.; Garcia-Villalba, R.; Mira, A.; Espin, J.C.; Tomas-Barberan, F.A. Isolation of human intestinal bacteria capable of producing the bioactive metabolite isourolithin A from ellagic acid. Front. Microbiol. 2017, 8, 1521. [Google Scholar] [CrossRef] [Green Version]
- Gaya, P.; Peirotén, Á.; Medina, M.; Álvarez, I.; Landete, J.M. Bifidobacterium pseudocatenulatum INIA P815: The first bacterium able to produce urolithins A and B from ellagic acid. J. Funct. Foods 2018, 45, 95–99. [Google Scholar] [CrossRef]
- Humphries, R.; Bobenchik April, M.; Hindler Janet, A.; Schuetz Audrey, N.; McAdam Alexander, J. Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, e00213–e00221. [Google Scholar] [CrossRef]
- Li, X.; Fields, F.R.; Ho, M.; Marshall-Hudson, A.; Gross, R.; Casser, M.E.; Naito, M. Safety assessment of Streptococcus salivarius DB-B5 as a probiotic candidate for oral health. Food Chem. Toxicol. 2021, 153, 112277. [Google Scholar] [CrossRef]
- Khatri, I.; Sharma, S.; Ramya, T.N.; Subramanian, S. Complete genomes of Bacillus coagulans S-lac and Bacillus subtilis TO-A JPC, two phylogenetically distinct probiotics. PLoS ONE 2016, 11, e0156745. [Google Scholar] [CrossRef]
- Landete, J.M. Ellagitannins, ellagic acid and their derived metabolites: A review about source, metabolism, functions and health. Food Res. Int. 2011, 44, 1150–1160. [Google Scholar] [CrossRef]
- González-Sarrías, A.; García-Villalba, R.; Núñez-Sánchez, M.Á.; Tomé-Carneiro, J.; Zafrilla, P.; Mulero, J.; Tomás-Barberán, F.A.; Espín, J.C. Identifying the limits for ellagic acid bioavailability: A crossover pharmacokinetic study in healthy volunteers after consumption of pomegranate extracts. J. Funct. Foods 2015, 19, 225–235. [Google Scholar] [CrossRef]
- Evtyugin, D.D.; Magina, S.; Evtuguin, D.V. Recent advances in the production and applications of ellagic acid and its derivatives. A review. Molecules 2020, 25, 2745. [Google Scholar] [CrossRef]
- Djedjibegovic, J.; Marjanovic, A.; Panieri, E.; Saso, L. Ellagic acid-derived urolithins as modulators of oxidative stress. Oxidative Med. Cell. Longev. 2020, 2020, 5194508. [Google Scholar] [CrossRef] [PubMed]
- Piwowarski, J.P.; Granica, S.; Kiss, A.K. Influence of gut microbiota-derived ellagitannins’ metabolites urolithins on pro-inflammatory activities of human neutrophils. Planta Med. 2014, 80, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Al-Harbi, S.A.; Abdulrahman, A.O.; Zamzami, M.A.; Khan, M.I. Urolithins: The gut based polyphenol metabolites of ellagitannins in cancer prevention, a review. Front. Nutr. 2021, 8, 647582. [Google Scholar] [CrossRef] [PubMed]
- Liou, C.S.; Sirk, S.J.; Diaz, C.A.C.; Klein, A.P.; Fischer, C.R.; Higginbottom, S.K.; Erez, A.; Donia, M.S.; Sonnenburg, J.L.; Sattely, E.S. A metabolic pathway for activation of dietary glucosinolates by a human gut symbiont. Cell 2020, 180, 717–728. [Google Scholar] [CrossRef]
- Clavel, T.; Lippman, R.; Gavini, F.; Dore, J.; Blaut, M. Clostridium saccharogumia sp. nov. and Lactonifactor longoviformis gen. nov., sp. nov., two novel human faecal bacteria involved in the conversion of the dietary phytoestrogen secoisolariciresinol diglucoside. Syst. Appl. Microbiol. 2007, 30, 16–26. [Google Scholar] [CrossRef]
- Hanske, L.; Loh, G.; Sczesny, S.; Blaut, M.; Braune, A. The bioavailability of apigenin-7-glucoside is influenced by human intestinal microbiota in rats. J. Nutr. 2009, 139, 1095–1102. [Google Scholar] [CrossRef]
- Selma, M.V.; Espin, J.C.; Tomas-Barberan, F.A. Interaction between phenolics and gut microbiota: Role in human health. J. Agric. Food Chem. 2009, 57, 6485–6501. [Google Scholar] [CrossRef] [PubMed]
- García-Villalba, R.; Giménez-Bastida, J.A.; Cortés-Martín, A.; Ávila-Gálvez, M.Á.; Tomás-Barberán, F.A.; Selma, M.V.; Espín, J.C.; González-Sarrías, A. Urolithins: A comprehensive update on their metabolism, bioactivity, and associated gut microbiota. Mol. Nutr. Food Res. 2022, 2022, e2101019. [Google Scholar] [CrossRef] [PubMed]
- Alegria, A.; Alvarez-Martin, P.; Sacristan, N.; Fernandez, E.; Delgado, S.; Mayo, B. Diversity and evolution of the microbial populations during manufacture and ripening of Casín, a traditional Spanish, starter-free cheese made from cow’s milk. Int. J. Food Microbiol. 2009, 136, 44–51. [Google Scholar] [CrossRef]
- Vendrell, D.; Balcazar, J.L.; Ruiz-Zarzuela, I.; De Blas, I.; Girones, O.; Muzquiz, J.L. Lactococcus garvieae in fish: A review. Comp. Immunol. Microbiol. Infect. Dis. 2006, 29, 177–198. [Google Scholar] [CrossRef]
- Shintani, M. The behavior of mobile genetic elements (MGEs) in different environments. Biosci. Biotechnol. Biochem. 2017, 81, 854–862. [Google Scholar] [CrossRef]
- Gibello, A.; Galan-Sanchez, F.; Blanco, M.M.; Rodriguez-Iglesias, M.; Dominguez, L.; Fernandez-Garayzabal, J.F. The zoonotic potential of Lactococcus garvieae: An overview on microbiology, epidemiology, virulence factors and relationship with its presence in foods. Res. Vet. Sci. 2016, 109, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Thurlow, L.R.; Thomas, V.C.; Fleming, S.D.; Hancock, L.E. Enterococcus faecalis capsular polysaccharide serotypes C and D and their contributions to host innate immune evasion. Infect. Immun. 2009, 77, 5551–5557. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Xie, J.; Zhang, M.; Fu, N.; Zhang, Y. Effect of a potential probiotics Lactococcus garvieae B301 on the growth performance, immune parameters and caecum microflora of broiler chickens. J. Anim. Physiol. Anim. Nutr. 2016, 100, 413–421. [Google Scholar] [CrossRef]
- Ayyash, M.; Abu-Jdayil, B.; Itsaranuwat, P.; Almazrouei, N.; Galiwango, E.; Esposito, G.; Hunashal, Y.; Hamed, F.; Najjar, Z. Exopolysaccharide produced by the potential probiotic Lactococcus garvieae C47: Structural characteristics, rheological properties, bioactivities and impact on fermented camel milk. Food Chem. 2020, 333, 127418. [Google Scholar] [CrossRef] [PubMed]
- Erkkilä, S.; Petäjä, E. Screening of commercial meat starter cultures at low pH and in the presence of bile salts for potential probiotic use. Meat Sci. 2000, 55, 297–300. [Google Scholar] [CrossRef]
- Ayyash, M.M.; Abdalla, A.K.; AlKalbani, N.S.; Baig, M.A.; Turner, M.S.; Liu, S.Q.; Shah, N.P. Invited review: Characterization of new probiotics from dairy and nondairy products-insights into acid tolerance, bile metabolism and tolerance, and adhesion capability. J. Dairy Sci. 2021, 104, 8363–8379. [Google Scholar] [CrossRef]
- Unban, K.; Kochasee, P.; Shetty, K.; Khanongnuch, C. Tannin-tolerant and extracellular tannase producing Bacillus isolated from traditional fermented tea leaves and their probiotic functional properties. Foods 2020, 9, 490. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Lyu, L.; Wang, Y.; Zhang, Y.; Guo, X.; Chen, Q.; Liu, C. Safety assessment and probiotic characteristics of Enterococcus lactis JDM1. Microb. Pathog. 2022, 163, 105380. [Google Scholar] [CrossRef]
- Tokatlı, M.; Gülgör, G.; Bağder Elmacı, S.; Arslankoz İşleyen, N.; Özçelik, F. In vitro properties of potential probiotic indigenous lactic acid bacteria originating from traditional pickles. BioMed Res. Int. 2015, 2015, 315819. [Google Scholar] [CrossRef]
- Liu, C.J.; Wang, R.; Gong, F.M.; Liu, X.F.; Zheng, H.J.; Luo, Y.Y.; Li, X.R. Complete genome sequences and comparative genome analysis of Lactobacillus plantarum strain 5-2 isolated from fermented soybean. Genomics 2015, 106, 404–411. [Google Scholar] [CrossRef]


| Antibiotic | Drug Contents (μg) | Standard for Judging Diameter of Inhibition Zone Diam (mm) [22] | Zone Diam (mm) | Antibacterial Effect | ||
|---|---|---|---|---|---|---|
| Resistant (R) | Intermediate (I) | Susceptible (S) | ||||
| Amikacin | 30 | ≤14 | 15~16 | ≥17 | 19 ± 0.5 | S |
| Norfloxacin | 10 | ≤12 | 13~16 | ≥17 | 12 ± 0.6 | R |
| Ofloxacin | 5 | ≤12 | 13~15 | ≥16 | 20 ± 0.3 | S |
| Ciprofloxacin | 5 | ≤15 | 16~20 | ≥21 | 17 ± 0.2 | I |
| Levofloxacin | 5 | ≤12 | 13~16 | ≥17 | 20 ± 0.1 | S |
| Erythromycin | 15 | ≤13 | 14~22 | ≥23 | 26 ± 0.6 | S |
| Tetracycline | 30 | ≤14 | 15~18 | ≥19 | 8 ± 0.3 | R |
| Cefuroxime | 30 | ≤14 | 15~17 | ≥18 | 35 ± 0.7 | S |
| Cefazolin | 30 | ≤14 | - | ≥15 | 31 ± 0.3 | S |
| Cefalotin | 30 | ≤14 | 15~17 | ≥18 | 25 ± 0.6 | S |
| Cefotaxime | 30 | ≤22 | 23~25 | ≥26 | 36 ± 0.2 | S |
| Cefatriaxone | 30 | ≤13 | 14~20 | ≥21 | 32 ± 0.2 | S |
| Ceftazidime | 30 | ≤14 | 15~17 | ≥18 | 30 ± 0.6 | S |
| Piperazoline | 100 | ≤28 | - | ≥29 | 31 ± 0.2 | S |
| Ampicillin | 10 | ≤16 | 18~24 | ≥25 | 29 ± 0.6 | S |
| Oxacillin | 1 | ≤17 | - | ≥25 | 17 ± 0.1 | R |
| Penicillin G | 10 | ≤28 | - | ≥29 | 33 ± 0.3 | S |
| Aztreonam | 30 | ≤15 | 16~21 | ≥22 | 0 | R |
| Co-trimoxazole | 23.75 | ≤10 | 11~15 | ≥16 | 0 | R |
| Furadantin | 300 | ≤14 | 15~16 | ≥17 | 23 ± 0.6 | S |
| Chloramphenicol | 30 | ≤12 | 13~17 | ≥18 | 26 ± 0.6 | S |
| BacillosporinB | 300 | ≤11 | 12~14 | ≥15 | 0 | R |
| Clindamycin | 2 | ≤13 | 14~17 | ≥18 | 0 | R |
| Kanamycin | 30 | ≤12 | 13~14 | ≥15 | 21 ± 0.2 | S |
| Gentamicin | 10 | ≤12 | 13~14 | ≥15 | 17 ± 0.3 | S |
| Streptomycin | 10 | ≤11 | 12~14 | ≥15 | 17 ± 0.5 | S |
| Vancomycin | 30 | ≤14 | 15~16 | ≥17 | 18 ± 0.2 | S |
| Resistance Type | Antibiotic Resistance | Identity (%) | Gene Locus |
|---|---|---|---|
| Tets | Tetracycline | 100 | GM_000049 |
| Vanra | Vancomycin, teicoplanin | 42.4 | GM_000289 |
| Pmra | Ciprofloxacin, norfloxacin | 50.0 | GM_000376 |
| Mdr | - | 41.2 | GM_000399 |
| Pbp2x | Penicillin | 42.7 | GM_000615 |
| Vanra | Vancomycin, teicoplanin | 43.7 | GM_000618 |
| Tet38 | Tetracycline | 42.8 | GM_000769 |
| Lsa | Lincosamide, streptogramin_b,Macrolide | 53.5 | GM_000802 |
| Emea | Fluoroquinolone | 82.0 | GM_000980 |
| Baca | Bacitracin | 56.9 | GM_001070 |
| Vanrg | Vancomycin | 46.4 | GM_001619 |
| Vanz | Teicoplanin | 45.5 | GM_001828 |
| Role | Virulence Factor | Related Genes | Identity (%) | Gene Locus |
|---|---|---|---|---|
| Adherence | Streptococcal plasmin receptor/GAPDH | plr/gapA | 84.8 | GM_001975 |
| EF-Tu | tuf | 72.6 | GM_001667 | |
| GroEL | groEL | 69.8 | GM_000286 | |
| Fibronectin-binding proteins | pavA | 63.3 | GM_000679 | |
| Immune modulation | Capsule | rmlC | 89.1 | GM_000129 |
| Capsule | rmlA | 88.9 | GM_000128 | |
| Capsule | STER_1222 | 85.6 | GM_000131 | |
| Capsule | hasC | 78.6 | GM_000658 | |
| Capsule | rmlD | 71.2 | GM_000132 | |
| Capsule | gnd | 68.2 | GM_001520 | |
| Capsule | STER_1434 | 62.9 | GM_000136 | |
| Exoenzyme | Streptococcal enolase | eno | 92.0 | GM_001501 |
| Hyaluronidase | EF0818 | 60.8 | GM_001436 | |
| Stress survival | Trigger factor | tig/ropA | 67.2 | GM_000359 |
| ClpP | clpP | 64.2 | GM_000379 | |
| ClpE | clpE | 62.5 | GM_001645 | |
| Regulation | LisR/LisK | lisR | 64.1 | GM_000453 |
| Type of Stress Response | Protein | Related Genes | Gene Locus |
|---|---|---|---|
| Acid stress response | F0F1-ATPase | atpA, atpB, atpC, atpD, atpE, atpF, atpG, atpH | GM_000429, GM_000430, GM_000431, GM_000432, GM_000433, GM_000434, GM_000435, GM_000436 |
| Na+/H+ antiporter family | - | GM_001303 | |
| Bile salts stress response | Cyclopropane-fatty-acyl-phospholipid synthase | cfa | GM_000663 |
| ABC transporter ATP-binding protein YxdL | yxdL | GM_000471 | |
| Sodium/hydrogen exchanger family | - | GM_000272, GM_000959, GM_000988, GM_001719 | |
| Temperature stress response | Heat shock protein 9/12 | - | GM_000598 |
| Cold shock protein | cspA | GM_000105, GM_000573, GM_001587, GM_001590 | |
| Metal stress response | Divalent metal cation transporter MntH | mntH | GM_000991 |
| Cation transport protein | - | GM_000063, GM_001371, GM_001582 | |
| CorA-like Mg2+ transporter protein | - | GM_001262, GM_001495, GM_001671, GM_001771 | |
| Cadmium, zinc and cobalt-transporting ATPase | cadA | GM_000577 | |
| Potassium/sodium uptake protein | ntpJ | GM_001371, GM_001582, GM_000063 | |
| Citrate-sodium symporter | citP | GM_001096 | |
| Oxidative stress response | Alkyl hydroperoxide reductase | ahpC | GM_001062 |
| Glutathione peroxidase | gpo | GM_000931 | |
| Thioredoxin reductases | trxB | GM_000780 | |
| Superoxide dismutase [Fe] | sodA | GM_000304 |
| Protein/Domain | Related Genes | Gene Locus |
|---|---|---|
| Segregation and condensation protein B | scpB | GM_001136 |
| Segregation and condensation protein A | scpA | GM_001137 |
| Flagellar hook-associated protein | flgK, flgL | GM_000659, GM_000034 |
| Laminin domain II | - | GM_001909 |
| Collagen binding domain | - | GM_001621 |
| Sortase A | strA | GM_000696 |
| S-ribosylhomocysteine lyase | luxS | GM_001854 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mi, H.; Liu, S.; Hai, Y.; Yang, G.; Lu, J.; He, F.; Zhao, Y.; Xia, M.; Hou, X.; Fang, Y. Lactococcus garvieae FUA009, a Novel Intestinal Bacterium Capable of Producing the Bioactive Metabolite Urolithin A from Ellagic Acid. Foods 2022, 11, 2621. https://doi.org/10.3390/foods11172621
Mi H, Liu S, Hai Y, Yang G, Lu J, He F, Zhao Y, Xia M, Hou X, Fang Y. Lactococcus garvieae FUA009, a Novel Intestinal Bacterium Capable of Producing the Bioactive Metabolite Urolithin A from Ellagic Acid. Foods. 2022; 11(17):2621. https://doi.org/10.3390/foods11172621
Chicago/Turabian StyleMi, Haoyu, Shu Liu, Yang Hai, Guang Yang, Jing Lu, Fuxiang He, Yaling Zhao, Mengjie Xia, Xiaoyue Hou, and Yaowei Fang. 2022. "Lactococcus garvieae FUA009, a Novel Intestinal Bacterium Capable of Producing the Bioactive Metabolite Urolithin A from Ellagic Acid" Foods 11, no. 17: 2621. https://doi.org/10.3390/foods11172621
APA StyleMi, H., Liu, S., Hai, Y., Yang, G., Lu, J., He, F., Zhao, Y., Xia, M., Hou, X., & Fang, Y. (2022). Lactococcus garvieae FUA009, a Novel Intestinal Bacterium Capable of Producing the Bioactive Metabolite Urolithin A from Ellagic Acid. Foods, 11(17), 2621. https://doi.org/10.3390/foods11172621






