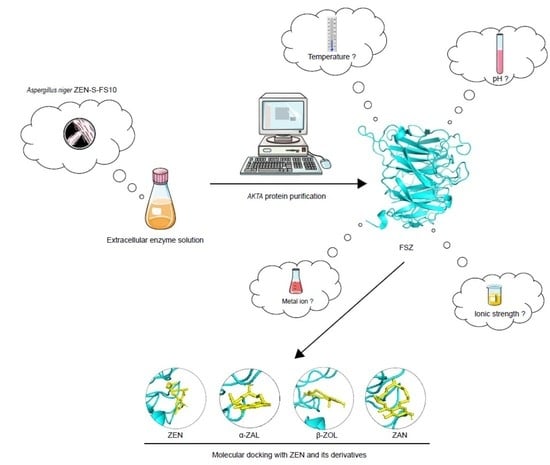
Isolation and Mechanistic Characterization of a Novel Zearalenone-Degrading Enzyme
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation of ZEN-S-FS10 and Its Crude Enzyme Solution
2.2. Purification of FSZ by ÄKTA Protein Purifier and SDS-PAGE Gel Electrophoresis
2.3. Detection of Enzyme Concentration and ZEN Degradation Activity
2.4. Determination of the Optimum Active Conditions for FSZ
2.4.1. Determination of Temperature Effects
2.4.2. Determination of pH Effects
2.4.3. Influence of Metal Ion
2.4.4. Influence of Ionic Strength
2.5. Determination of the Kinetic Parameters of ZEN Degradation by FSZ
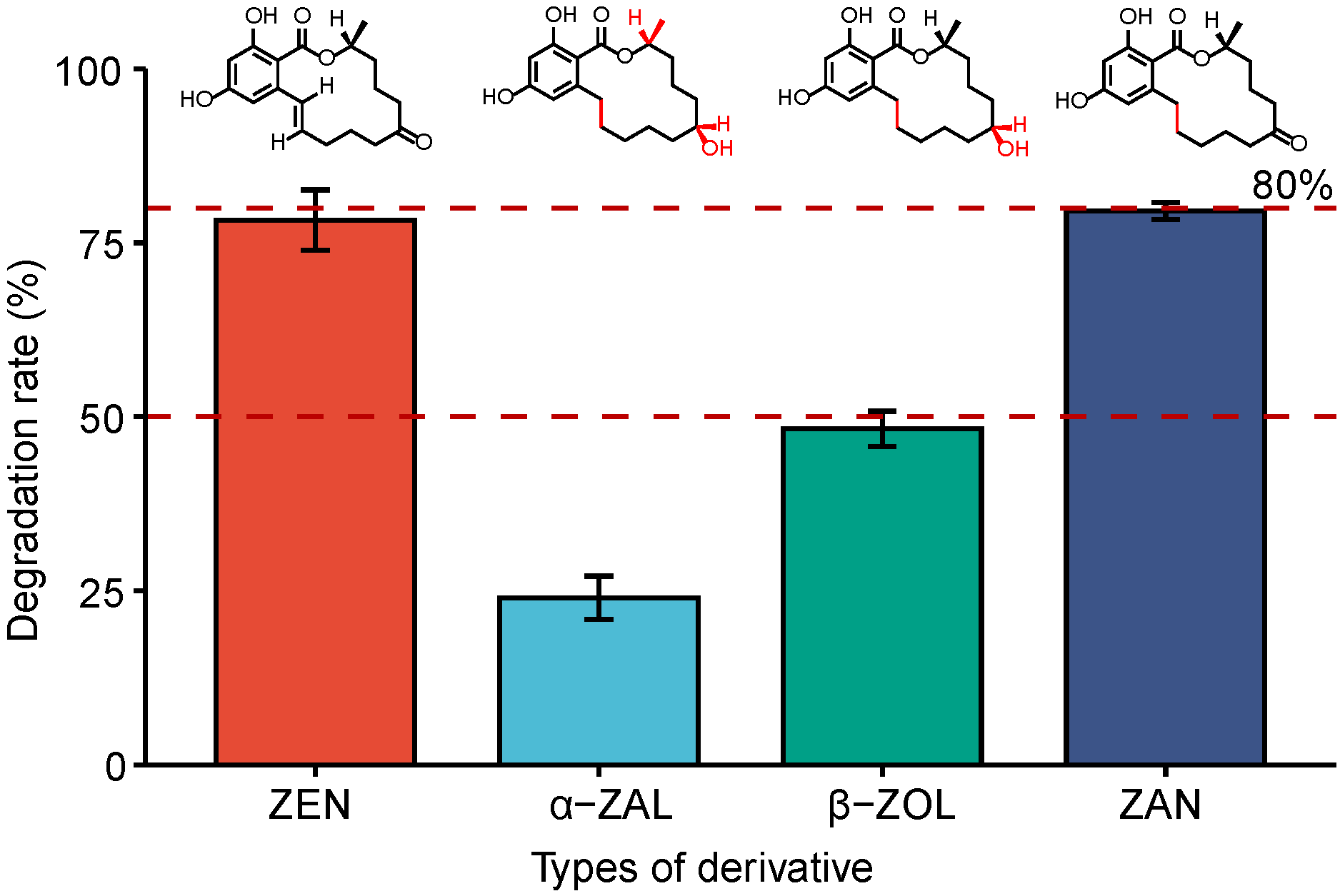
2.6. Measurement of Ability of FSZ to Degrade ZEN Derivatives
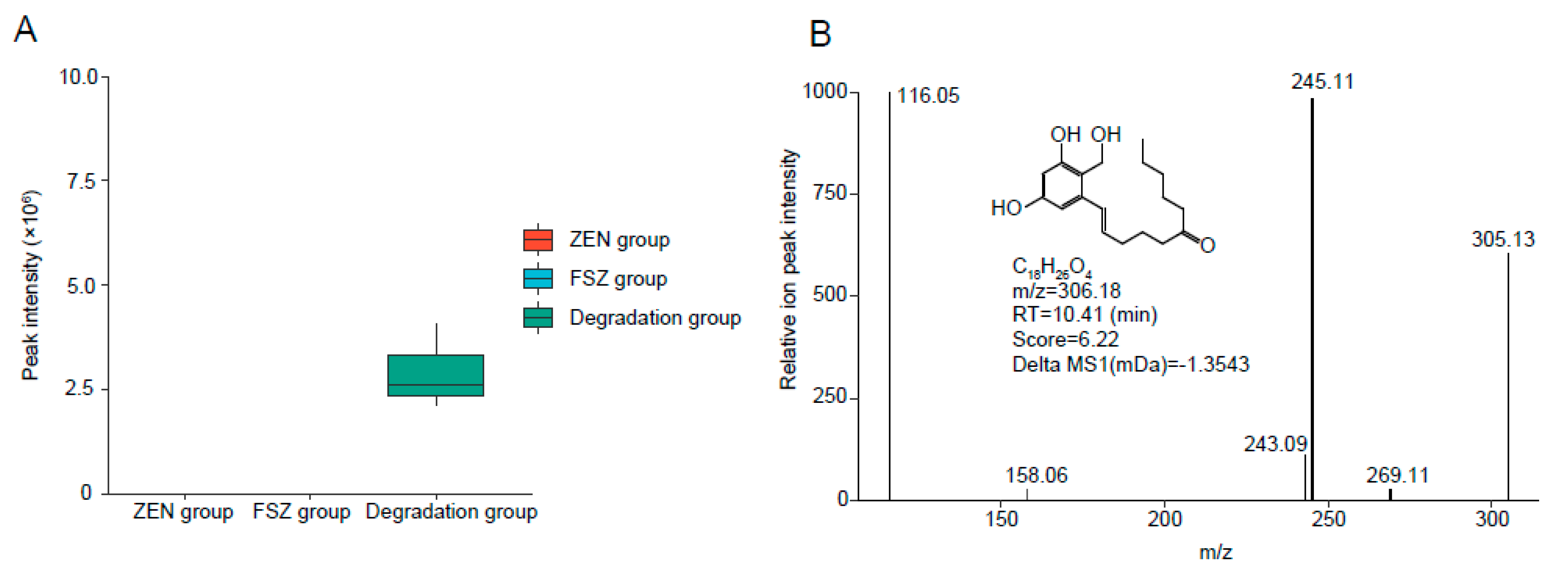
2.7. Exploration of ZEN Degradation Products
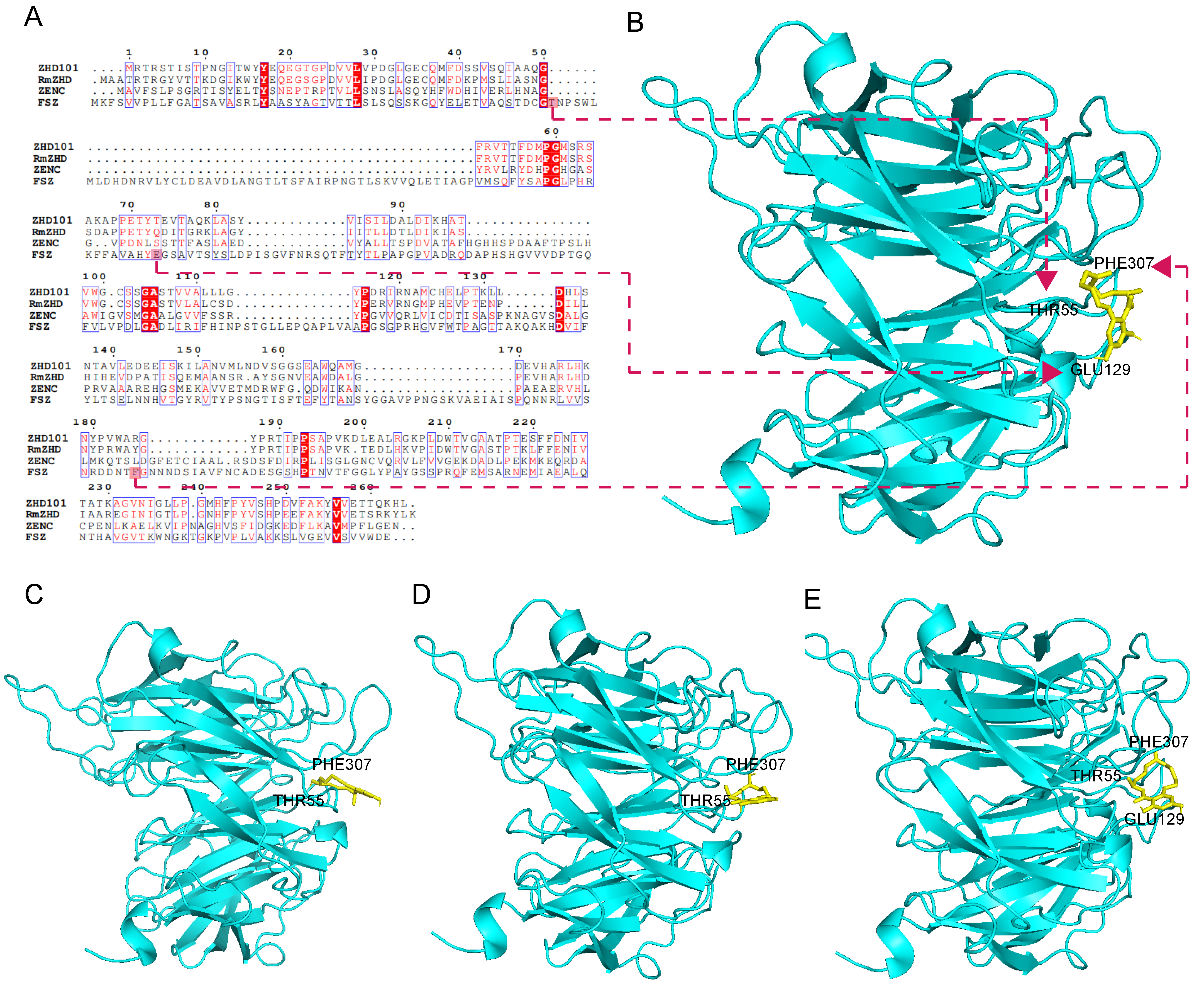
2.8. Sequence Analysis and Structural Characterization of FSZ
2.9. Evaluation of the Ability of FSZ to Degrade ZEN in Corn Flour
3. Results and Discussion
3.1. ÄKTA Protein Purification of FSZ
3.1.1. Purification of FSZ from Crude Enzyme Solution
3.1.2. Protein Concentration and Enzyme Activity after Purification
3.1.3. Degradability of Protein after Separation and Purification
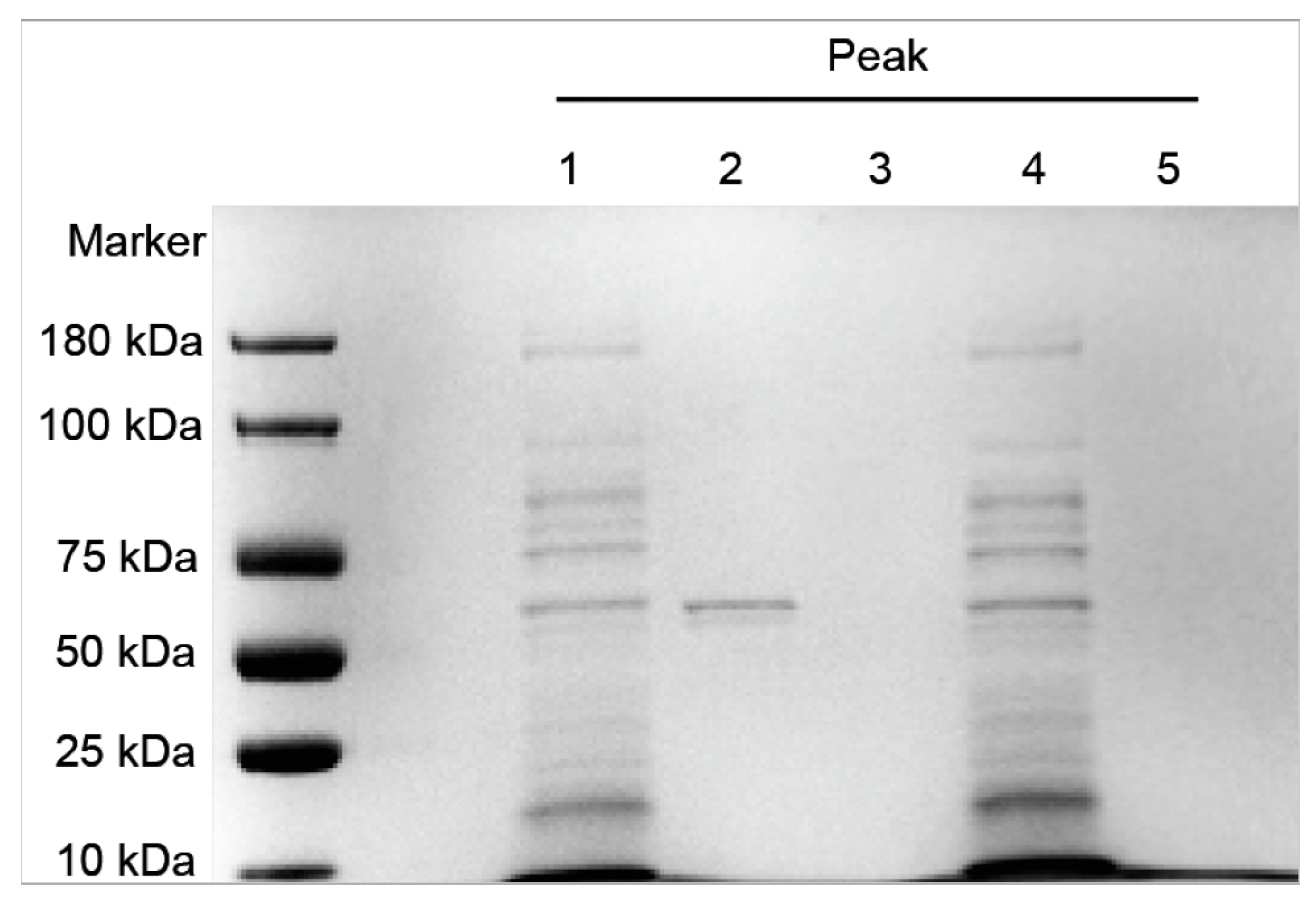
3.2. SDS-PAGE Analysis
3.3. Determination of the Optimal Active Conditions for FSZ
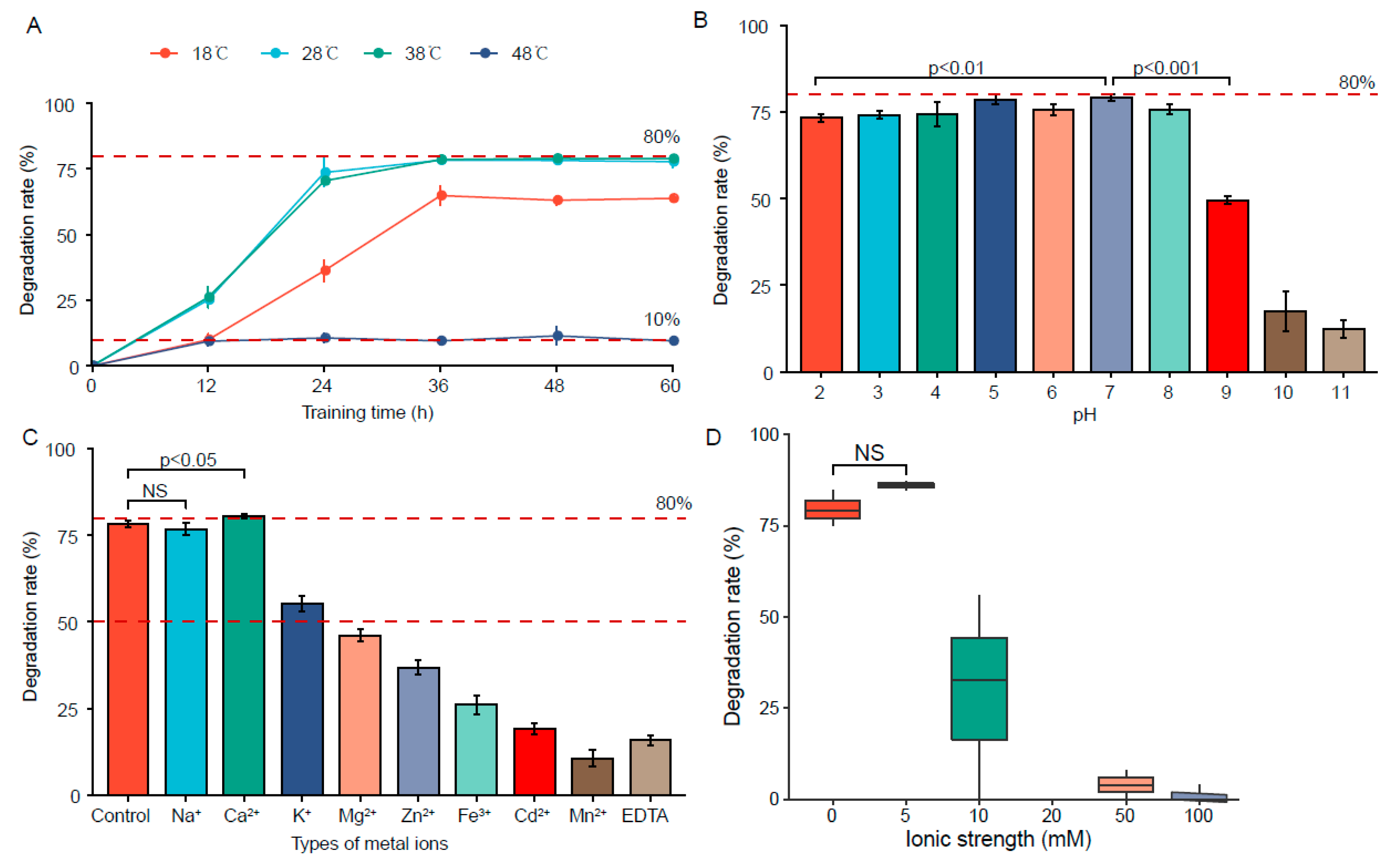
3.3.1. Effect of Temperature
3.3.2. Effect of pH
3.3.3. Effect of Metal Ions
3.3.4. Effect of Ionic Strength
3.4. Degradation Kinetic Analysis and Parameter Determination
3.5. Degradability of FSZ to ZEN Derivatives
3.6. Analysis of Products after FSZ Degradation of ZEN
3.7. Sequence Homology Comparison and Structural Analysis of FSZ
3.8. The Ability of FSZ to Degrade ZEN in Corn Flour
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stob, M.; Baldwin, R.S.; Tuite, J.; Andrews, F.N.; Gillette, K.G. Isolation of an Anabolic, Uterotrophic Compound from Corn Infected with Gibberella Zeae. Nature 1962, 196, 1318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wei, X.; Hao, W.; Wenli, Z.; Wanmeng, M. Identification of a Potent Enzyme for the Detoxification of Zearalenone. J. Agric. Food Chem. 2020, 68, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.; Borzekowski, A.; Haase, H.; Menzel, R.; Rueß, L.; Koch, M. Toxicity Assay for Citrinin, Zearalenone and Zearalenone-14-Sulfate Using the Nematode Caenorhabditis Elegans as Model Organism. Toxins 2018, 10, 284. [Google Scholar] [CrossRef] [PubMed]
- Faisal, Z.; Voros, V.; Fliszar-Nyul, E.; Lemli, B.; Kunsagi-Mate, S.; Poor, M. Interactions of Zearalanone, Alpha-Zearalanol, Beta-Zearalanol, Zearalenone-14-Sulfate, and Zearalenone-14-Glucoside with Serum Albumin. Mycotoxin Res. 2020, 36, 389–397. [Google Scholar] [CrossRef]
- Kakeya, H.; Takahashi-Ando, N.; Kimura, M.; Onose, R.; Yamaguchi, I.; Osada, H. Biotransformation of the Mycotoxin, Zearalenone, to a Non-Estrogenic Compound by a Fungal Strain of Clonostachys sp. Biosci. Biotechnol. Biochem. 2002, 66, 2723–2726. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Garcia, S.; Sibaja, K.V.M.; Nogueira, W.V.; Feltrin, A.C.P.; Pinheiro, D.F.A.; Cerqueira, M.B.R.; Furlong, E.B.; Garda-Buffon, J. Peroxidase as a Simultaneous Degradation Agent of Ochratoxin a and Zearalenone Applied to Model Solution and Beer. Food Res. Int. 2020, 131, 109039. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhu, L.D.; Chen, J.F.; Wang, W.; Zhang, R.M.; Li, Y.W.; Zhang, Q.Z.; Wang, W.X. Degradation Mechanism for Zearalenone Ring-Cleavage by Zearalenone Hydrolase Rmzhd: A Qm/Mm Study. Sci. Total Environ. 2020, 709, 10. [Google Scholar] [CrossRef]
- Takahashi-Ando, N.; Kimura, M.; Kakeya, H.; Osada, H.; Yamaguchi, I. A Novel Lactonohydrolase Responsible for the Detoxification of Zearalenone: Enzyme Purification and Gene Cloning. Biochem. J. 2002, 365, 1–6. [Google Scholar] [CrossRef]
- Bi, K.; Zhang, W.; Xiao, Z.H.; Zhang, D.W. Characterization, Expression and Application of a Zearalenone Degrading Enzyme from Neurospora Crassa. Amb. Express 2018, 8, 10. [Google Scholar] [CrossRef]
- Chang, X.; Liu, H.; Sun, J.; Wang, J.; Zhao, C.; Zhang, W.; Zhang, J.; Sun, C. Zearalenone Removal from Corn Oil by an Enzymatic Strategy. Toxins 2020, 1, 203. [Google Scholar] [CrossRef] [Green Version]
- Jun, W.; Xuewei, L.; Fangbo, Y.; Zhongbo, W.; Liuyan, Y. Cloning of a Dibutyl Phthalate Hydrolase Gene from Acinetobacter Sp. Strain M673 and Functional Analysis of Its Expression Product in Escherichia Coli. Appl. Microbiol. Biotechnol. 2013, 97, 2483–2491. [Google Scholar]
- Sun, X.; He, X.; Xue, K.s.; Li, Y.; Xu, D.; Qian, H. Biological Detoxification of Zearalenone by Aspergillus Niger Strain Fs10. Food Chem. Toxicol. 2014, 72, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Tianyu, Q.; Haiming, W.; Yang, Y.; Jian, Y.; Jian, J.; Jiadi, S.; Shuang, Z.; Xiulan, S. Exploration of Biodegradation Mechanism by Afb1-Degrading Strain Aspergillus Niger Fs10 and Its Metabolic Feedback. Food Control 2021, 121, 107609. [Google Scholar]
- Ji, J.; Yu, J.; Yang, Y.; Yuan, X.; Yang, J.; Zhang, Y.; Sun, J.; Sun, X. Exploration on the Enhancement of Detoxification Ability of Zearalenone and Its Degradation Products of Aspergillus Niger Fs10 under Directional Stress of Zearalenone. Toxins 2021, 13, 720. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.Z.; Zhang, Y.M.; Liu, Y.Y.; Zhang, M.; Lin, L.B.; Zhang, Q.L. Purification, Characterization, and Antibacterial and Antibiofilm Activity of a Novel Bacteriocin against Salmonella Enteritidis. Food Control 2021, 127, 10. [Google Scholar] [CrossRef]
- Zhu, Y.; Luo, G.; Li, Z.; Zhang, P.; Zhang, W.; Mu, W. Efficient Biosynthesis of Lacto-N-Neotetraose by a Novel Β-1,4-Galactosyltransferase from Aggregatibacter Actinomycetemcomitans Num4039. Enzym. Microb. Technol. 2022, 153, 109912. [Google Scholar] [CrossRef]
- Wu, S.; Wang, F.; Li, Q.; Wang, J.; Zhou, Y.; Duan, N.; Niazi, S.; Wang, Z. Photocatalysis and Degradation Products Identification of Deoxynivalenol in Wheat Using Upconversion Nanoparticles@Tio(2) Composite. Food Chem. 2020, 323, 126823. [Google Scholar] [CrossRef]
- David, K. Foodstuffs—Determination of Zearalenone and Trichothecenes Including Deoxynivalenol and Its Acetylated Derivatives (3-Acetyl-Deoxynivalenol and 15-Acetyl-Deoxynivalenol), Nivalenol T-2 Toxin and Ht-2 Toxin in Cereals and Cereal Products by Lc-Ms/Ms; European Committee for Standardization: Brussels, Belgium, 2019. [Google Scholar]
- Zhou, Z.; Luo, M.; Chen, X.; Yin, Y.; Xiong, X.; Wang, R.; Zhu, Z. Ion Mobility Collision Cross-Section Atlas for Known and Unknown Metabolite Annotation in Untargeted Metabolomics. Nat. Commun. 2020, 11, 4334. [Google Scholar] [CrossRef]
- De Mey, M.; Lequeux, G.J.; Maertens, J.; de Muynck, C.I.; Soetaert, W.K.; Vandamme, E.J. Comparison of Protein Quantification and Extraction Methods Suitable for E-Coli Cultures. Biologicals 2008, 36, 198–202. [Google Scholar] [CrossRef]
- Xiang, Y.Z.; Li, X.Y.; Zheng, H.L.; Chen, J.Y.; Lin, L.B.; Zhang, Q.L. Purification and Antibacterial Properties of a Novel Bacteriocin against Escherichia Coli from Bacillus Subtilis Isolated from Blueberry Ferments. Lwt-Food Sci. Technol. 2021, 146, 111456. [Google Scholar] [CrossRef]
- Butre, C.I.; Sforza, S.; Gruppen, H.; Wierenga, P.A. Introducing Enzyme Selectivity: A Quantitative Parameter to Describe Enzymatic Protein Hydrolysis. Anal. Bioanal. Chem. 2014, 406, 5827–5841. [Google Scholar] [CrossRef] [PubMed]
- Hollander, C.; Walrond, S.C.; Connelly, C.; Megson, L.; Bove, E.; McDonagh, T. A Custom Akta Avant Configuration Enabling Automated Parallel Protein Purification over a Range of Process Scales. Protein Expr. Purif. 2021, 182, 105842. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.O.; Feltrin, A.C.P.; Garda-Buffon, J. Zearalenone Reduction by Commercial Peroxidase Enzyme and Peroxidases from Soybean Bran and Rice Bran. Food Addit. Contam. Part A-Chem. Anal. Control Expo. Risk Assess. 2018, 35, 1819–1831. [Google Scholar] [CrossRef]
- Kosawang, C.; Karlsson, M.; Vélëz, H.; Rasmussen, P.H.; Collinge, D.B.; Jensen, B.; Jensen, D.F. Zearalenone Detoxification by Zearalenone Hydrolase Is Important for the Antagonistic Ability of Clonostachys Rosea against Mycotoxigenic Fusarium Graminearum. Fungal Biol. 2014, 118, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Urry, W.H.; Wehrmeister, H.L.; Hodge, E.B.; Hidy, P.H. The Structure of Zearalenone. Tetrahedron Lett. 1966, 7, 3109–3114. [Google Scholar] [CrossRef]
- Utermark, J.; Karlovsky, P. Role of Zearalenone Lactonase in Protection of Gliocladium Roseum from Fungitoxic Effects of the Mycotoxin Zearalenone. Appl. Environ. Microbiol. 2007, 73, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Castellano, F.N.; He, Z.W.; Greenaway, F.T. Hydroxyl Radical Production in the Reactions of Copper-Containing Amine Oxidases with Substrates. Biochim. Biophys. Acta 1993, 1157, 162–166. [Google Scholar] [CrossRef]
- Arif, T.; Currie, M.J.; Dobson, R.C.J.; Newson, H.L.; Poonthiyil, V.; Fairbanks, A.J.; North, R.A.; Rendle, P.M. Synthesis of N-Acetylmannosamine-6-Phosphate Derivatives to Investigate the Mechanism of N-Acetylmannosamine-6-Phosphate 2-Epimerase. Carbohydr. Res. 2021, 510, 108445. [Google Scholar] [CrossRef]
- Xiao, Z.C.; Hou, X.H.; Zhang, T.; Yuan, Y.H.; Xiao, J.B.; Song, W.; Yue, T.L. Starch-Digesting Product Analysis Based on the Hydrophilic Interaction Liquid Chromatography Coupled Mass Spectrometry Method to Evaluate the Inhibition of Flavonoids on Pancreatic Alpha-Amylase. Food Chem. 2022, 372, 131175. [Google Scholar] [CrossRef]
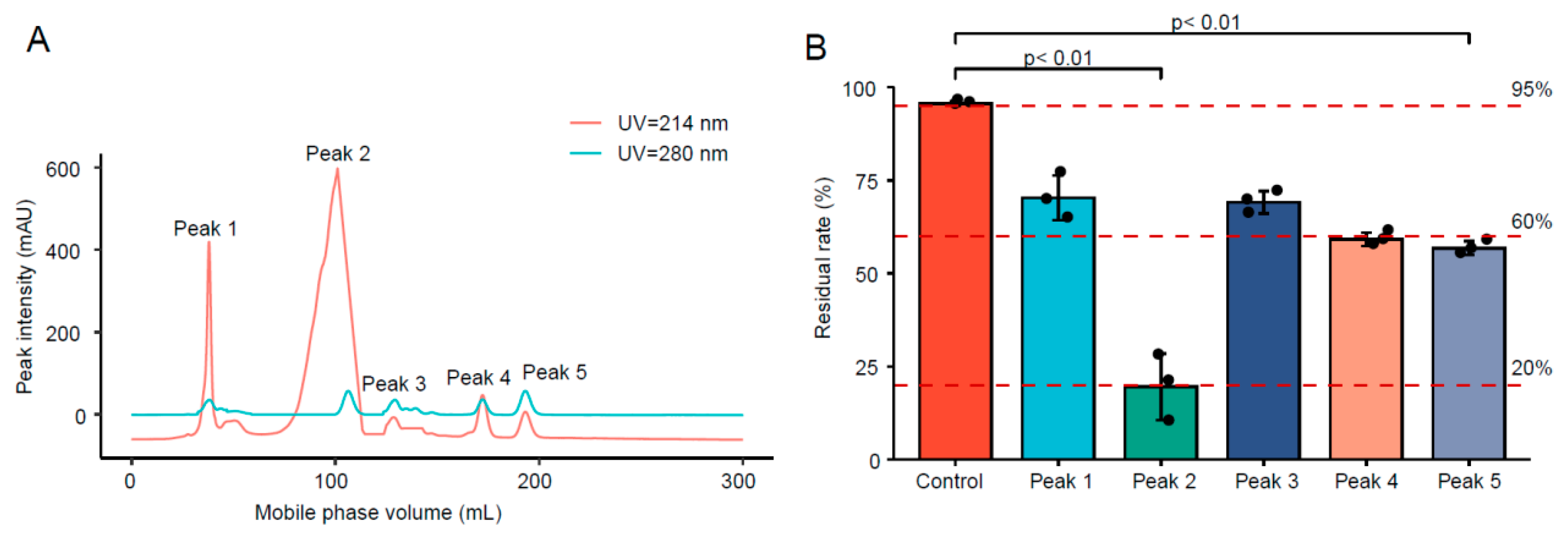

| Peak | Concentration (μg/mL) | Enzyme Activity (U/mg) |
|---|---|---|
| 1 | 6023.67 ± 5.09 | 5.82 ± 0.11 |
| 2 | 5887.67 ± 4.08 | 5.98 ± 0.05 |
| 3 | 5299.67 ± 6.14 | 2.11 ± 0.12 |
| 4 | 5116.63 ± 8.05 | 4.13 ± 0.02 |
| 5 | 5860.00 ± 2.59 | 3.12 ± 0.07 |
| ZEN-Degrading Enzymes | Degradation Rate (%) (24 h) | Vmax (μg·mL−1·h−1) | Km (μg/mL) | Source |
|---|---|---|---|---|
| FSZ | 75–80 | 6.52 | 0.85 | Aspergillus niger |
| POD [6] | 64.9 | 2.39 | 0.56 | - |
| Peroxidase [24] | 69.4 | 1.9 | 0.16 | Soybean bran |
| ZHD101 [25] | 50 | - | - | Clonostachys rosea |
| Analytical Parameters | ZEN |
|---|---|
| Curve equation | y = 0.0052x + 0.0062 |
| R2 | 0.9997 |
| LDm (μg/mL) | 0.05 |
| LQm (μg/mL) | 0.1 |
| Recovery rate (%) | 98.75 ± 1.11 |
| Degradation rate in corn flour (%) | 78.43 ± 1.78 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, J.; Yu, J.; Xu, W.; Zheng, Y.; Zhang, Y.; Sun, X. Isolation and Mechanistic Characterization of a Novel Zearalenone-Degrading Enzyme. Foods 2022, 11, 2908. https://doi.org/10.3390/foods11182908
Ji J, Yu J, Xu W, Zheng Y, Zhang Y, Sun X. Isolation and Mechanistic Characterization of a Novel Zearalenone-Degrading Enzyme. Foods. 2022; 11(18):2908. https://doi.org/10.3390/foods11182908
Chicago/Turabian StyleJi, Jian, Jian Yu, Wei Xu, Yi Zheng, Yinzhi Zhang, and Xiulan Sun. 2022. "Isolation and Mechanistic Characterization of a Novel Zearalenone-Degrading Enzyme" Foods 11, no. 18: 2908. https://doi.org/10.3390/foods11182908