1. Introduction
Kombucha is a traditional beverage of Asian origin, made by fermenting sugared green or black tea with a symbiotic culture of bacteria and yeasts within a cellulosic matrix. It has a long history of production and consumption that traces back to 220 BCE during the Tsin Dynasty in China, when it was considered a beverage with detoxifying and energizing properties [
1]. In recent years, kombucha consumption has grown in North America and Europe due to its refreshing taste and associated beneficial health properties which include, among others, antioxidant, antitumoral, anti-inflammatory, and hepatoprotective effects, digestion improvement, and microbial infection prevention [
1]. Even though these have yet to be fully proven in human clinical trials, these claims have contributed to the high popularity of kombucha [
2].
The microbial consortium responsible for tea fermentation is commonly named SCOBY (Symbiotic Culture of Bacteria and Yeasts) and is composed by acetic acid bacteria (AAB), yeasts, and lactic acid bacteria (LAB) [
3]. These microorganisms are embedded in an extracellular cellulosic matrix located at the liquid–air interphase. Several studies of kombucha fermentations have shown different microbial compositions of the SCOBY, which can vary depending on substrate, origin, climate, geographic location, and production conditions [
2,
4]. The most abundant yeasts present in the symbiotic culture belonged to the genera
Brettanomyces/Dekkera and
Zygosaccharomyces, while some reports also found
Saccharomyces,
Schizosaccharomyces, and
Hanseniaspora, among others. Regarding AAB,
Komagataeibacter,
Novacetimonas (a recently described genus including a group of former
Komagataeibacter spp. [
5]),
Acetobacter, and
Gluconobacter were prevalent, while among LAB, although not always present,
Oenococcus and
Liquorilactobacillus (former
Lactobacillus spp. [
6]) were the dominant genera [
3,
7].
The scientific exploration of this fermented beverage is quite difficult due to the huge diversity and abundance of microorganisms involved in its production, as well as the metabolic interactions between them. Nevertheless, some members of the consortium have a well-defined role in cooperative metabolism between the species, contributing to the chemical composition of kombucha. In fact, at the beginning of the fermentation process, the yeast community hydrolyzes sucrose into glucose and fructose by periplasmatic invertase and produces ethanol; this compound is metabolized by AAB to acetic acid, conferring the characteristic acidic aroma and flavor of vinegar to kombucha [
8]. Glucose and fructose are also used by AAB to produce cellulose, leading to the formation of a cellulosic matrix [
8]. Particularly, some species, belonging to the genera
Komagataeibacter and
Novacetimonas, such as
Komagataeibacter xylinus,
Novacetimonas hansenii, and
Novacetimonas maltaceti, were found to be very efficient producers of this biopolymer [
5,
9]. This matrix is an adhesion surface and protects the embedded cells from unfavorable environmental factors, such as ultraviolet radiation or heat [
8]. Furthermore, it exposes the AAB to the aerobic environment, which is essential for their growth and metabolism. The role of LAB has been mainly linked to their ability to produce lactic and gluconic acids that enhance the antimicrobial and antioxidant properties of kombucha—and could also impact the overall sensory perception [
10]. These bacteria were reported to comprise up to 30% of the bacterial population of kombucha cultures [
2], even though several surveys have not detected their presence at all [
3,
11,
12].
The chemical composition of kombucha has been largely described in literature [
13,
14], revealing that some compounds, such as organic acids, vitamins, polyphenols, and amino acids, are present at high concentrations [
15]. However, ample variations in composition and metabolite concentration have been observed, as they depend on several factors, such as type of tea, fermentation time, and microorganisms of the inoculum [
16]. Indeed, several laboratory fermentations of kombucha using different SCOBY consortia and tea brews led to a broad range of beverages that differed in terms of their chemical composition, including the relative amount of ethanol and acetic acid [
7,
17]. Some studies also analyzed the volatile organic compounds (VOCs) of kombucha, which, in addition to confirming variability between different batches, revealed that acids and esters were the most important contributors to overall organoleptic properties, as they were associated with the common descriptors of the product, e.g., acidic, refreshing, and possessing a fruity aroma [
9,
10]. Quality attributes related to VOCs have defined taste panel protocols for sensory analysis, selecting appropriate descriptors (i.e., sweetness, sourness, bitterness) [
18]. Kombucha tea taste is generally described by evaluators as pleasantly acidic and harmonic, with a fresh, sour, fruity, and vinegar-like flavor [
19,
20].
High reproducibility and control of the fermentation process help to ensure a better quality of the final product, but the complex nature of the SCOBY traditionally used to start new fermentations makes it difficult to standardize Kombucha production. Indeed, the production of batches by successive propagation could lead to the evolution of the complex microbial consortium in terms of composition, microbial dynamics, or both [
21]. The fluctuation of biological components could lead to unstable qualities of kombucha fermentation kinetics and chemical composition and, consequently, organoleptic properties [
22]. To overcome this, it is necessary to understand the role of each microbial component of SCOBY, depicting its contribution to the final organoleptic characteristics of the beverage, and selecting the most significant species and strains to simplify kombucha consortia. Indeed, a limited number of strains with desirable characteristics, such as
B. bruxellensis and
K. rhaeticus strains, can produce a kombucha-like fermentation without other taxa [
10,
23].
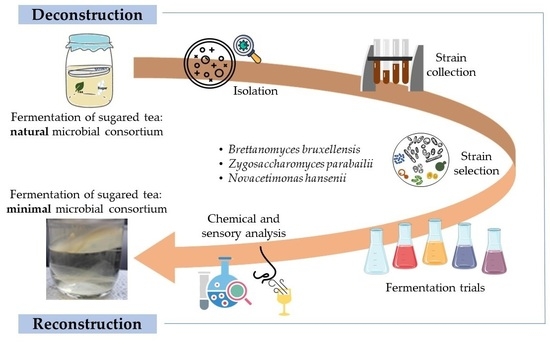
In this context, the present study aimed to investigate the microbial structure of an artisanal kombucha, with the ultimate goal of establishing a collection of taxonomically well-defined strains to be used for an ad hoc consortium development. Adopting a microbiota deconstruction–reconstruction approach in a controlled laboratory environment, the dominant and peculiar bacteria and yeast strains were selected, and their contribution to the aromatic profile and organic acid production was explored. The derived tailor-made microbial community could be used to standardize the production process and the quality of the final product.
2. Materials and Methods
2.1. Analyzed Samples, Microbial Enumeration and Isolation
Samples were collected during the manufacture of kombucha at an artisanal producer located in Verona, Italy. The fermented beverage was prepared by infusing a 0.3% w/v blend of green and black tea (80% and 20%, respectively) with 5% w/v brown sugar, and a kombucha culture from a previous batch (20% v/v liquid phase and 0.28% w/v SCOBY), which was used to inoculate microorganisms and decrease the pH below 4.6. The fermentation was carried out in a dedicated room with controlled temperature (28 ± 1 °C), to ensure better growth conditions and to limit the occurrence of possible contaminations. A total of 11 samples were collected in November 2020 from five tanks; three of them, i.e., T2 (21 days of fermentation), T3 and T4 (14 days), were used for production of kombucha and the other two (T1, T5) for SCOBY maintenance and propagation. SCOBY was maintained in a smaller tank (T1), submerged in tea, while for propagation, another tank (T5) with an increased liquid/air interface area was used. Samples were distinguished in SCOBY (seven samples) and liquid phase (four samples). All samples were transferred to the laboratory in refrigerated conditions and stored at 4 °C.
For microbiological analysis, SCOBY samples (5 g) were first resuspended in 5 mL physiological solution (0.9% w/v NaCl) and then disrupted with an ULTRA-TURRAX disperser (IKA-Werke, Staufen, Germany). Decimal dilutions in physiological solution of homogenized SCOBY and liquid phase samples were then plated on Wallerstein Lab (WL) agar medium (ThermoFisher Scientific, Waltham, MA, USA) supplemented with chloramphenicol (100 mg/L) for yeasts, MRS agar (Oxoid, Basingstoke, England, UK) for LAB, and GYC (50 g/L glucose, 10 g/L yeast extract, 30 g/L CaCO3, 25 g/L agar) supplemented with 100 mg/L cycloheximide and 100 mg/L nystatin for AAB. All media were incubated at 28 °C for up to 5 days. MRS plates were incubated anaerobically with AnaerocultTM A (Merck, Darmstadt, Germany). After counting, representative colonies were picked up and purified by successive streaking on their respective isolation media. Putative yeasts, AAB, and LAB isolates were routinely grown at 28 °C for three days in YPD (10 g/L yeast extract, 20 g/L bacteriological peptone, 20 g/L glucose), GY (50 g/L glucose, 10 g/L yeast extract), and MRS broth, respectively. Isolates were stored at −80 °C in 25% v/v glycerol (Thermo Scientific, Monza, Italy) solution. Unless otherwise specified, reagents were purchased from Sigma-Aldrich (Milan, Italy).
2.2. DNA Extraction and Taxonomic Identification
Total genomic DNA was obtained from a 2-mL aliquot of three-days-grown AAB, LAB, and yeast cultures. Briefly, cells were harvested by centrifugation (14,000 rpm for 10 min), washed with distilled water, and then resuspended in 10 mg/µL lysozyme solution (Sigma-Aldrich), for bacterial isolates, or 10 U/µL lyticase solution (Sigma-Aldrich), for yeasts. After incubation (37 °C, 1 h), DNA isolation followed the protocol of Wizard Genomic DNA Purification Kit (Promega, Milan, Italy). DNA yield and purity were determined with a NanoDrop ND1000 UV–Vis Spectrophotometer (Thermo Scientific, Waltham, MA, USA), and DNA samples were stored at −20 °C for downstream analysis.
Rep-PCR with primer (GTG)
5 was employed according to the protocol of [
24], modified by [
25,
26]. PCR amplification was conducted in a Thermal Cycler 2720 (Applied Biosystems, Foster City, CA, USA) with the following program for bacterial DNA: 5 min of denaturation at 94 °C, 30 cycles of 1 min at 95 °C, 1 min at 40 °C, and 8 min at 72 °C, then 16 min at 72 °C for the final elongation. As for yeast DNA, the program was modified as follows: 5 min at 94 °C, 35 cycles of 50 s at 95 °C, 50 s at 40 °C, and 90 s at 72 °C, then 5 min at 72 °C. The products were run on a 1.5%
w/
v agarose gel in 1× TAE buffer (40 mM Tris, 20 mM acetic acid, and 0.4 mM EDTA) stained with Atlas Clearsight (Bioatlas, Tartu, Estonia) at 110 V for 2 h. Each gel was loaded with the molecular ladder O’Gene Ruler DNA (Thermo Scientific) to normalize the runs.
Visualization and image capturing were made under UV light with the UVITEC Gel Documentation System (Cleaver Scientific, Rugby, England). Fingerprinting data were then analyzed using Bionumerics v.7.6. (Applied Maths, Sint-Martens-Latem, Belgium), with the Pearson’s correlation coefficient and the unweighted pair group method with arithmetical average (UPGMA) clustering method.
Identification of representative strains of different Rep-PCR clusters were done by 16S rRNA gene and 26S rRNA gene sequencing for LAB and yeasts, respectively. Identification of AAB were performed through
dnaK gene sequencing, due to the high similarity between 16S rRNA gene of different AAB species. The 16S rRNA gene was amplified using the primers E8F and E1541R, according to [
27], while the D1/D2 domain of the 26S rRNA gene was amplified using primers NL-1 and NL-4, following the protocol of [
28].
The gene
dnaK was amplified by the newly designed primers DNAKF (fwd-TGAAGTGCTGCGTATCATCAACGA) and DNAKR (rev-ATTTCACGTTCGCCCTGATA) with the following program: 5 min of denaturation at 95 °C, 30 cycles of 30 s at 95 °C, 30 s at 60 °C, and 1 min at 72 °C, then 10 min at 72 °C for the final elongation, using the following reagent concentrations: DreamTaq Green Buffer (Thermo Scientific) 1X, dNTPs 250 μM, primer DNAKF 1 μM, primer DNKAR 1 μM, Dream Taq (Thermo Scientific) 0.025 U/μL, DNA 50 ng/μL. The PCR products were purified and sent for sequencing at Eurofins Genomics (Ebersberg, Germany). Yeasts and AAB sequencing data were searched against the GenBank database using the BLAST alignment tool (
http://blast.ncbi.nlm.nih.gov/, accessed on 4 July 2022), while 16S rRNA gene sequencing data were searched against bacterial type strains 16S rRNA gene sequences in the EzBioCloud database (
https://www.ezbiocloud.net/, accessed on 4 July 2022). The sequencing data generated in this study were all deposited in the NCBI database within the projects n. BankIt2616791 for
dnaK gene sequences of AAB, SUB11972523 for 16S rRNA gene sequences of LAB, and SUB11972681 for 26S rRNA gene sequences of yeasts.
2.3. Fermentation Trials
Lab-scale fermentation trials were carried out with mono- and co-cultures of distinctive strains: B. bruxellensis T7SB-5W6, N. hansenii T7SS-4G1, and Z. parabailii T7SS-4W1. They were compared with the native microbial consortium used for kombucha manufacture by the artisanal producer. Sugared tea (2.5% w/v fructose and 2.5% w/v glucose) was prepared by infusing 0.3% w/v tea leaves (80% green and 20% black) in water at 85 °C for 5 min and acidifying with acetic acid (Sigma-Aldrich) to bring the pH below 4.6. To prepare the inoculum, cells were cultivated in the respective growth media, collected by centrifugation (5000 rpm, 5 min), washed twice with physiological solution, counted by plating method, and inoculated in tea. Regarding the native consortium, 40 mL of the artisanal kombucha was centrifuged and the obtained pellet was washed twice with physiological solution, counted, and inoculated in tea. For all trials, the initial microbial concentration was about 105 CFU/mL, determined by plate count after inoculum. The fermentation trials were carried out in triplicate in 200 mL volume and incubated at 28 °C for 14 days.
The different fermentation strategies were labeled with the following codes: KCC (native microbial consortium); TEA (non-inoculated sugared tea used as control for the chemical analysis); Nh (monoculture of N. hansenii T7SS-4G1); Zp (monoculture of Z. parabailii T7SS-4W1); Bb (monoculture of B. bruxellensis T7SB-5W6); NhZp (co-culture of N. hansenii T7SS-4G1 + Z. parabailii T7SS-4W1), NhBb (co-culture of N. hansenii T7SS-4G1 + B. bruxellensis T7SB-5W6); NhZpBb (reconstructed microbial consortium with N. hansenii T7SS-4G1 + Z. parabailii T7SS-4W1 + B. bruxellensis T7SB-5W6).
The pH was measured throughout the fermentation process with the pH-meter Crison Basic 20 (Hach-Lange, Barcelona, Spain). In samples inoculated with
N. hansenii T7SS-4G1, cellulosic matrix production was quantified at the end of fermentation by weighting the samples after water removal at 60 °C for 3 days [
29].
2.4. High-Performance Liquid Chromatography (HPLC)
To quantify sugars (glucose, fructose), organic acids (acetic, glucuronic, and gluconic acids), and alcohols (glycerol, ethanol) during kombucha tea trials, HPLC analysis was performed at 0, 7, and 14 days of fermentation using the Extrema LC-4000 system (Jasco, Cremella, Italy) coupled with a refractive index detector RI-4030 (Jasco) set to 35 °C. Analytes were separated using a RezexTM ROA-Organic Acid H + (8%) column (300 × 7.8 mm; Phenomenex, Castel Maggiore, Italy) equipped with a Carbo-H (4 × 3.0 mm) guard column (Phenomenex). The column was maintained at a constant temperature of 80 °C and under an isocratic mobile phase consisting of 5 mM H2SO4 (Honeywell, Rodano, Italy) at a flow rate of 0.8 mL/min. Before analysis, samples were centrifugated at 6000× g for 5 min, filtered with 0.22 µm syringe filters SPHEROS (LLG Labware, Meckenheim, Germany), and appropriately diluted with 5 mM H2SO4. Calibration curves were prepared in a range from 0.05 to 6 g/L.
2.5. Analysis of Volatile Compounds
For quantification of alcohols, esters, fatty acids, and benzenoids (except methyl salicylate), the procedure described by [
30] was followed, performing a Solid Phase Extraction (SPE) through a BOND ELUT-ENV cartridge (Agilent Technologies. Santa Clara, CA, USA); 50-mL samples of kombucha tea fermentation trials with addition of 100 µL internal standard (2-octanol, 4.2 mg/L in ethanol) were diluted with 50 mL of deionized water. Samples were loaded on the SPE cartridge, previously activated with 20 mL of dichloromethane, 20 mL of methanol, and equilibrated with 20 mL of water. After sample loading, the cartridges were washed with 15 mL of water. Volatile compounds were eluted with 10 mL of dichloromethane and then concentrated under gentle nitrogen stream to 200 μL prior to GC injection.
Free terpenes, norisoprenoids, and methyl salicylate were quantified using Solid Phase Micro Extraction (SPME), following the procedure described by [
31]; 5 mL of samples with addition of 5 µL internal standard (2-octanol, 4.2 mg/L in ethanol) were diluted with 5 mL of deionized water and placed into a 20-mL glass vial together with 3 g of NaCl. SPME extraction was performed using a 50/30 μm divinylbenzene–carboxen–polydimethylsiloxane (DVB/CAR/PDMS) fiber (Supelco, Bellafonte, PA, USA) exposed to sample headspace for 60 min at 40 °C.
Volatile sulfur-containing compounds were quantified according to [
32] by placing 10 mL of sample with 100 µL of internal standard (DMS-d6, 2 mg/L in ethanol) and 3 g of NaCl into a 20-mL glass vial. SPME extraction was performed using a polydimethylsiloxane-divinylbenzene fibre (PDMS/DVB) (Supelco, Bellafonte, PA, USA) exposed to sample headspace for 30 min.
Gas Chromatography-Mass Spectrometry (GC-MS) analysis was carried out on an HP 7890A (Agilent Technologies, Cernusco sul Naviglio, Italy) gas chromatograph coupled to a 5977B quadrupole mass spectrometer, equipped with a Gerstel MPS3 auto sampler (Müllheim an der Ruhr, Germany). Separation was performed using a DB-WAX UI capillary column (30 m × 0.25 mm, 0.25 μm film thickness, Agilent Technologies) and helium (6.0 grade) as carrier gas at 1.2 mL/min of constant flow rate. The GC oven was programmed as follows: started at 40 °C for 3 min, raised to 230 °C at 4 °C/min and maintained for 20 min. A different program of the GC oven was applied to the vials for quantification of volatile sulfur-containing compounds: started at 35 °C for 5 min, increased to 90 °C at 5 °C/min and then to 260 °C at 15 °C/min, maintained for 2 min. Mass spectrometer was operated in electron ionization (EI) at 70 eV with ion source temperature at 250 °C and quadrupole temperature at 150 °C. Mass spectra were acquired in Selected Ion Monitoring (SIM) mode. Samples were analyzed in random order.
Calibration curves were prepared for all quantification methods. For SPE-GC-MS method, a calibration curve was prepared for each analyte using seven concentration points and three replicates per point in model solution (2% v/v ethanol, 3.5 g/L tartaric acid, pH 3.0), 100 µL of internal standard 2-octanol (4.2 mg/L in ethanol) was added to each calibration solution, which was then submitted to SPE extraction and GC-MS analysis as described for the samples. For SPME-GC-MS method, a calibration curve was prepared for each analyte using seven concentration points and three replicate solutions per point in model solution. Five µL of internal standard 2-octanol (4.2 mg/L in ethanol) and 100 µL of DMS-d6 (2 mg/L in ethanol) were added to each calibration solution, which was then submitted to SPME extraction and GC-MS analysis as described for the samples. Calibration curves were obtained using Chemstation software (Agilent Technologies) by linear regression, plotting the response ratio (analyte peak area divided by internal standard peak area) against concentration ratio (added analyte concentration divided by internal standard).
2.6. Sensory Evaluation
Sensory evaluation of kombucha tea was carried out by means of the sorting task methodology, as described by [
33], with slight modifications. Sixteen panelists (eleven males and five females) participated in the sessions. One hour before the test, samples were removed from the 16 °C cold room and 20 mL of each fermentation trial were poured in ISO glasses (
https://www.iso.org/standard/9002.html, accessed on 23 August 2022), labeled with 3-digit random codes, and covered by plastic Petri dishes; all samples were served at 22 ± 1 °C, and glasses were presented in random order for each panelist. To assess the reproducibility of the panel, a replicate of one sample was added to the sorting task. Panelists were asked to sort the kombucha samples into groups based on odor similarities exclusively by orthonasal evaluation, with no request to indicate specific odor descriptors, and without limitations on group number.
2.7. Statistical Analysis
Data of analytical determinations on GC-MS were compared by three-way analysis of variance (ANOVA), followed by the post-hoc Tukey’s HSD (Honestly Significant Difference) test, with statistical significance threshold set at 95% (
p-value < 0.05), and Principal Component Analysis (PCA). GC-MS data were averaged, centered, and scaled by compound and hierarchically clustered by Ward’s minimum variance method and Euclidean distance metric with the hclust R [
34] function before being plotted by the heatmap.2 function. For the sorting task, data was organized into individual similarity binary matrices (8 × 8 samples; 0 = different and 1 = similar) for each panelist. The dendrogram of hierarchical cluster analysis with the Ward criterion was obtained from a co-occurrence matrix, calculated by the sum of all panelists. Statistical analysis was performed using the package XL-STAT (Addinsoft SARL, Paris, France).
4. Discussion
Kombucha is a fermented beverage resulting from the activity of a complex microbial consortium, in which the contributions of each component have only recently been investigated [
10,
20,
35,
36]. In the present study, a microbiota deconstruction/reconstruction approach, combined with analysis of biochemical parameters and fermentation metabolites with organoleptic impact, was applied to guide strain selection and standardize kombucha quality.
Initially, the ecological study carried out on the native microbial consortium of the artisanal kombucha tea allowed us to depict its structure, set up a strain collection, and select distinctive strains to develop an ad hoc consortium. Microbiological analysis highlighted the presence of a large number and diversity of microorganisms, confirming previous results [
17], with higher counts of both yeasts and bacteria in SCOBY samples, accounting its ability to protect and trap microorganisms [
33]. These data corresponded to those found by [
7], who reported, for cellulosic matrix, a minimal count of 7 Log CFU/g, and a lower value for liquid samples (4.4 Log CFU/g) [
37].
The molecular analysis of yeast isolates led to the identification of three species:
B. bruxellensis,
B. anomalus, and
Z. parabailii.
Brettanomyces has been described as the most dominant yeast genus in several black and green tea fermentations [
3,
7,
8]; it is well adapted to harsh environmental conditions, such as low pH and high ethanol concentrations. Generally,
B. anomalus is prevalent in several kombucha teas [
3,
7], but in the present study,
B. bruxellensis was found more abundantly. This aspect could have been related to the intraspecific variability of
B. bruxellensis in the acetic acid metabolism, which can enable specific strains to outcompete other microbes [
38]. This characteristic could be further investigated for the strain
B. bruxellensis T7SB-5W6, recovered in most of the analyzed samples.
Zygosaccharomyces has been frequently detected by culture-dependent and metagenomic analysis among the dominant genera in kombucha [
39,
40]. However, the species
Z. parabailii, isolated from the sample T2Sb, has been rarely found in this beverage as it was recovered only in one sample out of 16 samples of different brands and countries [
13]. As such, its contribution to kombucha characteristics has not yet been totally unraveled. Interestingly, in one study
Z. parabailii was found to be co-dominant with
B. bruxellensis in kombucha obtained with black, green, and rooibos tea [
17].
As for AAB, the genera
Komagataeibacter, Novacetimonas, and
Acetobacter have been commonly found in kombucha, also by culture-independent methods, revealing that these AAB are dominant and well-adapted to this matrix [
3,
12,
39]. In particular,
N. hansenii can represent around 20% of AAB in both black and green kombucha teas [
7], and is a species of industrial interest known for its application in bacterial cellulose production.
Liquorilactobacillus nagelii was the only LAB species with a unique strain detected in this study: a Gram-positive, rod-shaped facultative anaerobe that grows well in an atmosphere enriched with CO
2 [
41].
L. nagelii has been previously identified in kombucha samples [
7,
10], however, the strain
L. nagelii TLV-4R7 isolated in the present study was not included in the fermentation trials for its very slow growth rate in a preliminary evaluation.
The development of a collection of strains isolated from kombucha and their features was the starting point to design lab-scale tea fermentation trials using different combinations of the strains
N. hansenii T7SS-4G1,
Z. parabailii T7SS-4W1, and
B. bruxellensis T7SB-5W6. All microbial combinations caused a pH decrease, as previously observed in other kombucha fermentation studies [
12,
42], mainly due to glucose and fructose metabolism during microbial fermentation and related production of organic acids, especially acetic acid. This was dependent on the microbial population, substrate composition, and fermentation time [
36]. The rapid pH decrease in the first days was mainly associated with bacterial metabolism [
10], as seen in this study and confirmed by the slower decrease observed in yeast monocultures. Additionally, in accordance with the present investigation, previous reports showed a slower pH decrease toward the end of fermentation, with a stabilization around the 10th day thanks to a buffering effect in the fermented medium [
10,
43,
44].
Regarding the different starters inoculated, the Bb monoculture maintained the pH around 4.0 after 10 days of fermentation, which was comparable to the value of 4.17 obtained by [
45] after 12 days, even if the starting pH was very different (4.6 against 6.9, respectively). Nh fermentation pH after 10 days was 2.55 and remained stable until the 14th day, which was concordant with the pH value of 2.6 after 7 days found by [
46]. Those authors, who used a
N. hansenii strain isolated from kombucha, stated that a lower pH would inhibit microbial growth and bacterial cellulose production [
46]. The consortia KCC and NhZpBb showed a similar pH after 14 days of fermentation (2.58 and 2.60, respectively). These results agreed with [
22], who found a pH of 2.4 at the end of fermentation using a reconstructed consortium with
Acetobacter pasteurianus,
K. xylinus, and
Z. bailii. On the contrary, the minimal consortium with
Acetobacter indonesiensis,
Hanseniaspora valbyensis, and
B. bruxellensis proposed by [
20] reached a pH value of 4.17 at the end of fermentation, although starting from 6.9.
Sugar depletion was peculiar for each monoculture, reflecting species-specific attitudes.
Z. parabailii consumed exclusively fructose. This could be attributed to its fructophilic behavior, which was linked to the presence of high-capacity and low-affinity fructose transporters [
25].
B. bruxellensis also consumed a small amount of fructose, as most
Brettanomyces strains have the capability to metabolize a wide range of monosaccharides, disaccharides, trisaccharides, and dextrins [
38]. In [
12], a monoculture of
B. bruxellensis consumed around 10 g/L of total sugar after 14 days of fermentation, the same value found in the present Bb fermentation, while our strain of
Z. parabailii consumed two times that amount.
Notable differences in sugar depletion were observed between KCC and NhZpBb, possibly indicating that
Z. parabailii played a major role in sugar consumption when part of the reconstructed consortium, while its activity was not prevalent in the native microbial consortium. Contrarily, [
20] did not find noteworthy differences on the consumption of sugars when testing different strains in mono- and co-cultures to evaluate a proposed minimal consortium.
Chemical parameters, as cellulose, acetic acid, glycerol, and ethanol, were quantified during the fermentation trials. KCC showed significant lower production of those metabolites than NhZpBb, which could be correlated with the lower sugar consumption. In binary combinations, such as NhZp and NhBb, it became clear how yeast-bacterium interactions substantially influenced the chemical composition and were dependent of yeast species due to differential growth rates and metabolic pathways. Different microbial populations and interactions among them might produce kombucha with divergent fermentation kinetics and chemical outcomes, despite encountering the same conditions [
2].
The highest amount of cellulose was produced by Nh monoculture, possibly because there was no competition with other microorganisms, and glucose could be exploited to produce cellulosic matrix. As a matter of fact, the more complex consortia KCC and NhZpBb had the lowest yield of cellulose. Only the yeast monocultures could not produce any cellulose at all. No data are currently available for cellulose dry weight production, but [
47] reported, for cellulose wet weight, a yield of 3.25 g/g of L-sucrose, using a commercial kombucha as inoculum. In the study of [
12], a consistent cellulosic biofilm was only formed with a native kombucha consortium; in the co-cultures of selected strains, the biofilm was not present. Using two different yeast species,
Zygosaccharomyces bisporus and
B. bruxellensis, in pairwise combinations with
K. rhaeticus, the production of biofilm was remarkably higher with
B. bruxellensis than with
Z. bisporus [
23], thus opposite of this study where more cellulose was recovered from NhZp than NhBb.
The authors of [
7] reported acetic acid concentrations between of 7.65–9.18 g/L for green and black tea kombucha, while [
10] and [
20], investigating reconstructed consortia, found lower levels of 2.41 g/L and 1.05 g/L, respectively. In the present study, values ranged from 0.32 to 8.37 g/L, except for Nh. The lowest end, as expected, was represented by the yeast monocultures, due to the absence of bacterial conversion of ethanol to acetic acid, while the highest production was obtained with the reconstructed consortium NhZpBb.
Novacetimonas spp. dominate vinegar fermentation, where they are most used, thanks to the resistance to ethanol and acetic acid [
48]. Some AAB can use glucose to produce acetic acid, and this compound could be further oxidized via acetyl-CoA synthesis and the TCA cycle [
12,
22]. Hence, the absence of acetic acid in Nh could be explained by a lack of production or a later consumption of this compound due to oxidation reactions.
Ethanol and glycerol production differed significantly among fermentations, generally reaching higher levels in the presence of Zp, behavior that could be related to the fructophilic character of this species [
42,
43]. The monoculture Zp produced more ethanol than Bb, and the co-culture NhZp produced more than NhBb. In both co-cultures, a decrease of ethanol was observed as compared with the monocultures, associated with the production of acetic acid from ethanol promoted by Nh [
8]. A divergent result was found by [
23], who reported a strikingly higher ethanol production by
B. bruxellensis than
Z. bisporus.
In NhZpBb, more acetic acid and ethanol were produced than NhZp and NhBb. Possibly, the combined activity of the two yeasts generated more ethanol, thus more substrate was available for the bacterial metabolism to produce acetic acid, also as a response to the stress caused by other strains. Furthermore, acetic acid could stimulate the yeasts to produce more ethanol [
12,
22], resulting in both metabolites increasing. In previous studies with reconstructed kombucha consortia, other authors found lower ethanol levels than the present investigation, e.g., 0.237 g/L [
10] and 0.3 g/L [
22].
The ethanol conversion promoted by bacteria in the symbiotic cultures helps to decrease alcohol volume below the legal limits for the classification of non-alcoholic beverages, which is 0.5%
v/
v in the USA, Brazil, Australia, and New Zealand, and 1.2%
v/
v in the European Union [
10,
16]. Indeed, in the present investigation, both yeast monocultures exceeded limits, with 0.53%
v/
v in Bb and 1.24%
v/
v in Zp. Regarding co-cultures, these values dropped, and only the kombucha produced with the reconstructed consortium would have needed to be labeled as alcoholic in the first countries, reaching 0.89%
v/
v. Nevertheless, as reported by [
16], several commercial kombucha teas presented more than 0.5%
v/
v ethanol in the analytical determinations, although these were not correctly labeled.
In the case of glycerol, the dependence on the metabolism of
Z. parabailii for the generation of this compound was even clearer, as Zp, NhZp, and NhZpBb reached around 0.83 g/L after 14 days of fermentation, while the fermentations without
Z. parabailii were all below 0.10 g/L. Final concentration of glycerol was reported to be around 0.1–0.5 g/L [
9] and 2 g/L [
3] in fermented tea, comparable to the present study. Interestingly,
B. bruxellensis produced less than 1:10 in respect to
Z. parabailii, and the presence of
N. hansenii did not influence glycerol production. Glycerol production is normally associated with osmotolerant yeasts, such as the genera
Candida,
Pichia,
Schizosaccharomyces,
Torulaspora, and
Zygosaccharomyces [
3]. Some
Novacetimonas (former
Komagataeibacter) prefer the oxidation of ethanol, whereas
Gluconobacter in general favor the oxidation of glycerol, glucose, gluconic acid, and sorbitol over ethanol [
48]. Glycerol has been related to a positive influence on body and viscosity in wine and can increase the sweetness and smoothness of the beverage [
49].
Volatilome profiles depict each strain contribution to microbial consortia, linking chemical parameters to possible sensory repercussions. Alcohols (e.g., isoamyl alcohol, phenylethyl alcohol), fatty acids (e.g., hexanoic acid, octanoic acid), esters (e.g., ethyl acetate, isoamyl acetate), and terpenes (e.g., linalool, β-citronellol) production were well correlated with yeast activity (
Z. parabailii and
B. bruxellensis), as also reported in [
20]. Some molecules were also differentially produced between the two yeast species, since they could take different routes for nutrient utilization and production of secondary metabolites, even though central carbon metabolism was mostly conserved [
50]. Zp monoculture produced almost six times more isoamyl alcohol compared to Bb and three times more phenylethyl alcohol, confirmed in co-culture, in which the contribution of
Z. parabailii was crucial. Phenylethyl alcohol was found in greater proportion in kombucha with higher ethanol and glycerol [
3], which was also observed in the present study, associated with the presence of
Z. parabailii.
Fermentations with
B. bruxellensis produced the highest overall concentration of fatty acids, which could be related to the fact that other kombucha produced in the absence of this species had the lowest level of acids [
3]. Interestingly, Nh showed a comparable lauric acid concentration to Bb (105.50 and 130.00 µg/L, respectively) and the production was greatly enhanced when they were co-cultured, reaching 701.55 in NhBb and 742.50 µg/L in NhZpBb, but that was still less than half the amount reached in KCC sample (1472.00 µg/L). Lauric acid has a strong antimicrobial activity, as it is effective against bacteria that are prevalent in overweight people, thus helping to control obesity [
4]. In the case of isovaleric acid, the concentrations in all co-cultures were superior to those found in the monocultures, also highlighting the synergistic effects of the microbial interactions. Moreover, this molecule is commonly produced by
B. bruxellensis; however, it was coupled with unpleasant acidity in kombucha, and described as earthy, medicinal, and sweaty [
19,
20]. Fatty acids are generally released in wine during autolysis of yeasts and could be involved in the yeasty aroma of kombucha, and alongside higher alcohols, they promote aromas of vinegar, apple juice, and exotic fruits [
20].
Almost all esters, terpenes, and norisoprenoids reached their highest concentrations in the kombucha fermented with the consortia KCC and NhZpBb. Those are important aromatic compounds in fermented beverages, usually associated with pleasant floral and fruity aromas [
51,
52]. Esters enhance tea and white fruit odor perceptions in kombucha [
20]. Ethyl acetate production was enhanced when all strains were co-cultured, reaching 51.30 µg/L, while both pairwise yeast-bacteria combinations failed to reach more than 10 µg/L. This effect could be explained by yeast capacity of converting ethanol into ethyl acetate, where yeast synergy is required to obtain a higher amount of this compound [
53]. The synergistic effect was observed for the other esters as well, except for ethyl octanoate and phenethyl acetate, which were strongly associated with
B. bruxellensis and
Z. parabailii, respectively. The first also reached the highest concentration of the precursor octanoic acid, while the latter produced the most phenylethyl alcohol—as expected, considering the yeast metabolic pathways of fatty acid ethyl esters and acetate esters, respectively [
52].
Regarding terpenes, the synergy of multiple species increased their levels, but some individual contributions of the selected strains were clearly acknowledged.
B. bruxellensis could be associated with β-citronellol and
Z. parabailii with linalool. NhZpBb co-culture showed the highest linalool increase (11.27 µg/L), where the main yeast responsible was
Z. parabailii which in monoculture reached 8.17 µg/L, almost doubling the concentration found for
B. bruxellensis (4.14 µg/L). Linalool content increased in honey byproduct fermentation by a microbial consortium that included
Z. bailii [
54]. Although terpenes have been considered varietal aromas and
de novo biosynthesis of terpenes by
Saccharomyces and non-
Saccharomyces yeasts was normally neglected, some
S. cerevisiae and
Hanseniaspora uvarum wild strains produced up to 4 µg/L linalool and 1 µg/L β-citronellol in a chemically defined must-like medium [
55].
B. bruxellensis, in monoculture, produced the highest concentrations of the benzenoids 4-ethylphenol and methyl salicylate, while single
Z. parabailii was the biggest producer of the sulfur-containing compounds dimethyl sulfide and methionol, and benzaldehyde. Methyl salicylate (spicy, wintergreen) and benzaldehyde (cherry, almond), as linalool and β-citronellol, are mainly released from bound glycosides in fermented beverages by yeast-derived glycosidase enzymes, and they are all important contributors to the flavor of tea [
56,
57].
Brettanomyces spp. and
Zygosaccharomyces spp. were considered to have low β-glucosidase activity [
19], albeit a β-glucosidase isolated from
B. anomalus enhanced methyl salicylate and linalool in forest fruit milk [
58]. As for 4-ethylphenol, its formation from p-coumaric acid has been exhaustively described in
B. bruxellensis, but elevated concentrations can give aromas of horse sweat, barnyards, and medicine [
59]. Dimethyl sulfide and methionol, with aromas of cabbage, truffle, and boiled potato, are volatile sulfur-containing compounds produced during fermentation by yeast metabolism, highly dependent of species and strain. Concentrations of these two compounds were strikingly higher in Zp, but we did not find any study about volatile sulfur-containing compounds metabolism, either with
Zygosaccharomyces spp. or
Brettanomyces spp., despite extensive research with
S. cerevisiae and other non-conventional yeasts [
60,
61] To the best of our knowledge, this was the first study ever to consider the presence of terpenes, norisoprenoids, and sulfur-containing compounds in kombucha, even though these molecules and their precursors were already described in tea [
56].
The ability of
Z. parabailii to produce alcohols and sulfur-containing compounds was compressed in the reconstructed consortia, while the production of fatty acids and benzenoids by
B. bruxellensis was maintained or augmented when co-inoculated with the other strains. Accumulation of esters was led by yeast metabolism and not affected by the yeast-bacteria interaction, but a synergistic effect was seen in the consortium comprising the two yeast strains. For terpenes and norisoprenoids, the concentration in the reconstructed consortium was possibly a contribution of the three strains. These changes, associated with the microbial interactions, could be related with modifications to oxygen access due to pellicle formation, alterations of pH, competition for substrates, and cross-feeding of metabolites [
10,
45].
Sensory analysis was conducted to evaluate both mono- and co-cultures by sorting task method. The clustering was not identical to the VOCs profile, but some trends were still visible, and certain correlations could be suggested between the chemical analysis and the sensory outcomes. Peculiar VOCs production of Bb and Zp affected panelists’ evaluation, showing a distinctive profile from other samples. The separation of the monocultures from the other fermentations was most likely defined by the much lower concentration of acetic acid. The authors of [
36] defined, in kombucha, an odor perception threshold of 0.21 g/L for acetic acid, whereas it became pungent in concentrations above 1.25 g/L. Therefore, acetic acid could be theoretically perceived in all fermentations, but it only became pungent in the co-cultures and microbial consortia.
The two binary co-cultures clustered together in both analyses; however, in sensory analysis, they clustered closer to the bacterial monoculture than to the yeasts. This could suggest that, although yeast metabolism was more influent in the carbon metabolism and production of secondary metabolites, participation of the bacteria was more influential in the sensory response. The reconstructed consortium (NhZpBb) was the most closely related to KCC, possibly because these two samples had the highest concentrations of fatty acids, which can mask the aromatic notes of tea esters and ketones [
20]. Nevertheless, the presence of fatty acids precursors could contribute to the biosynthesis of their respective ethyl esters, thus modulating the aromatic equilibrium [
62]. Indeed, the higher levels of acetic, hexanoic, and octanoic acids in NhZpBb and KCC were accompanied by increased ethyl acetate, ethyl hexanoate, and ethyl octanoate.
These results showed that the combination of the three selected strains was enough to mimic a more complex kombucha consortium made up of more species, providing a possible way to standardize commercial kombucha production. Satisfactory products fermented by a new tailor-made consortium, with similarities to classical kombucha obtained by inoculation of the established consortium from a previous batch, were also observed in previous sensory analysis [
20,
36].