Inhibitory Mechanism of Baicalein on Acetylcholinesterase: Inhibitory Interaction, Conformational Change, and Computational Simulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Enzyme Activity Assay
2.3. Kinetic Analysis of the Inhibitory Type
2.4. Fluorescence Spectrum Measurement
2.5. CD Spectra Measurements
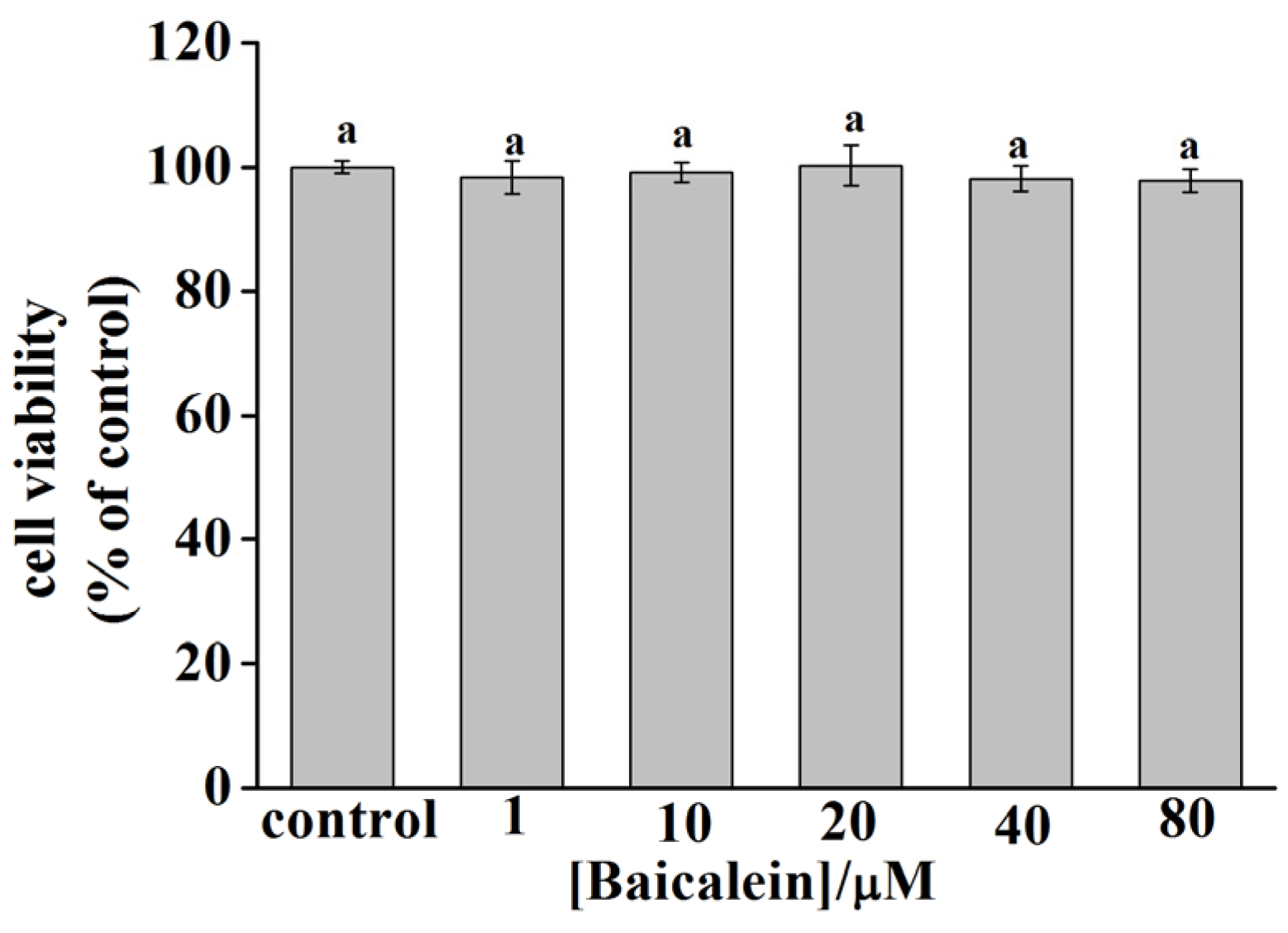
2.6. Cytotoxicity of Baicalein in PC12 Cells
2.7. Molecular Docking
2.8. M.D. Analysis
2.9. Statistical Analysis
3. Results and Discussion
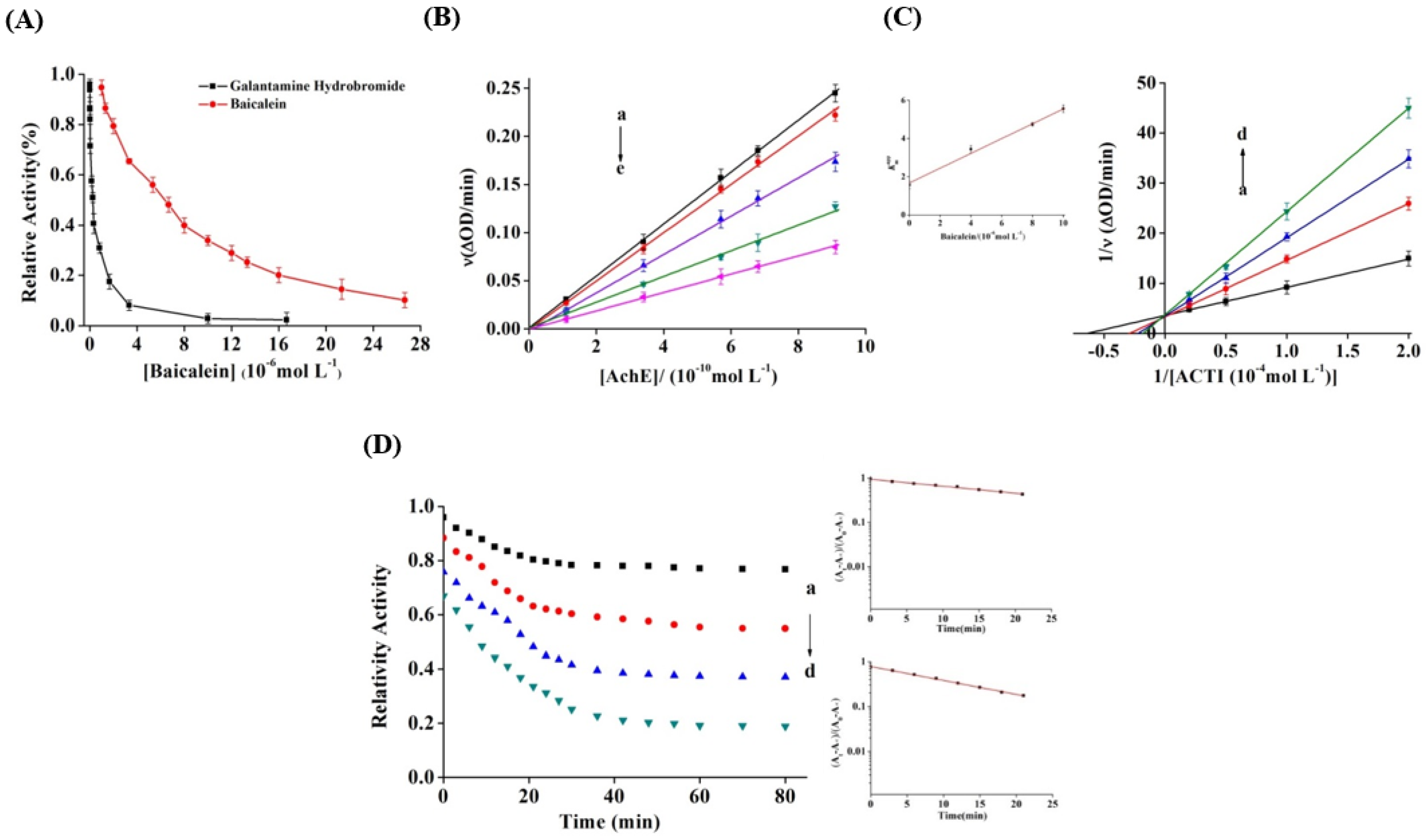
3.1. Inhibitory Effect of Baicalein on AChE
3.2. Inhibition Kinetics of Baicalein
3.3. Inactivation Kinetics and Rate Constants
3.4. Fluorescence Quenching of AChE by Baicalein
3.5. Binding Parameters and Thermodynamics
3.6. Synchronous Fluorescence Spectroscopy
3.7. CD Spectra
3.8. Cytotoxicity of Baicalein in PC12 Cells
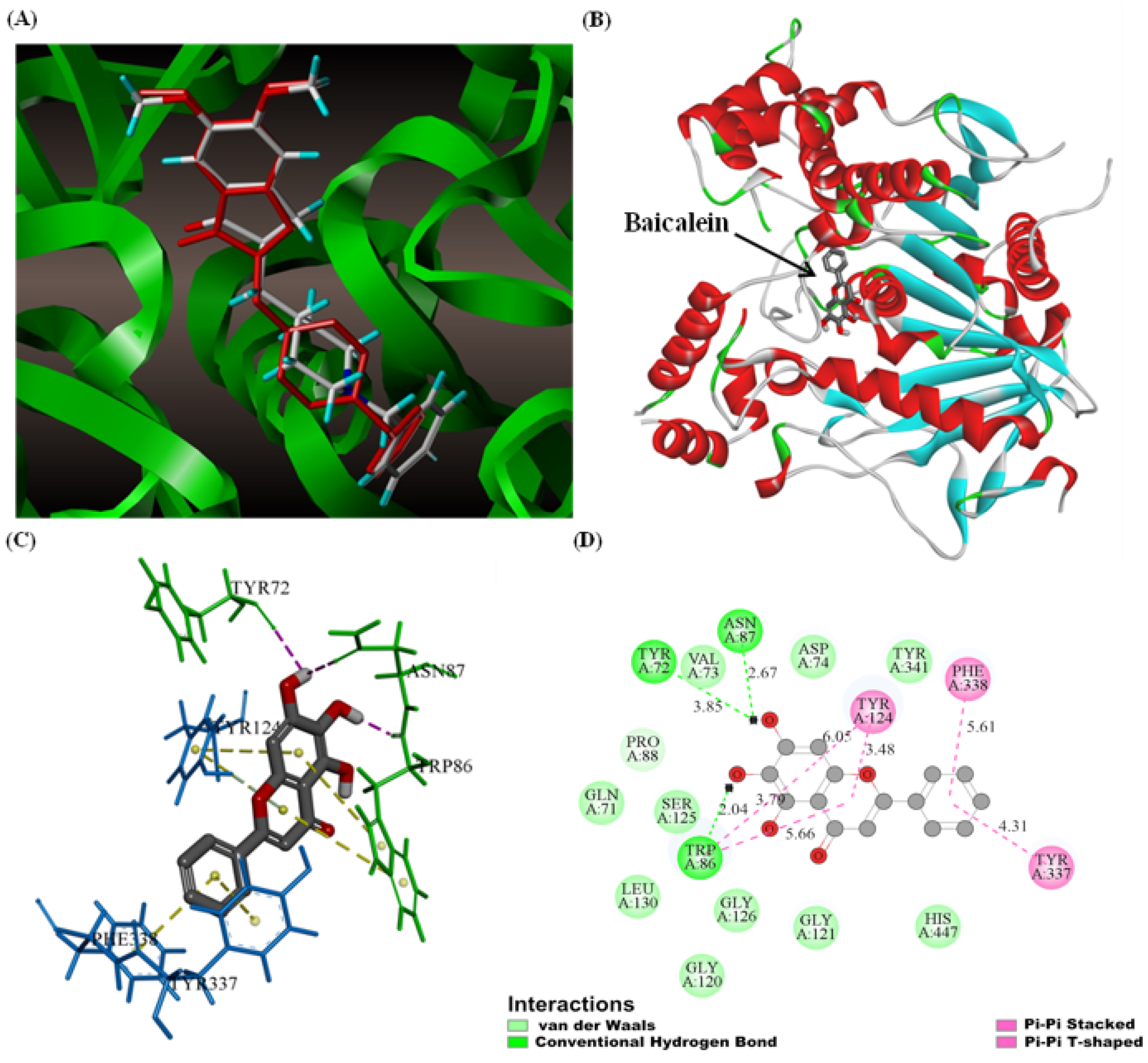
3.9. Molecular Docking
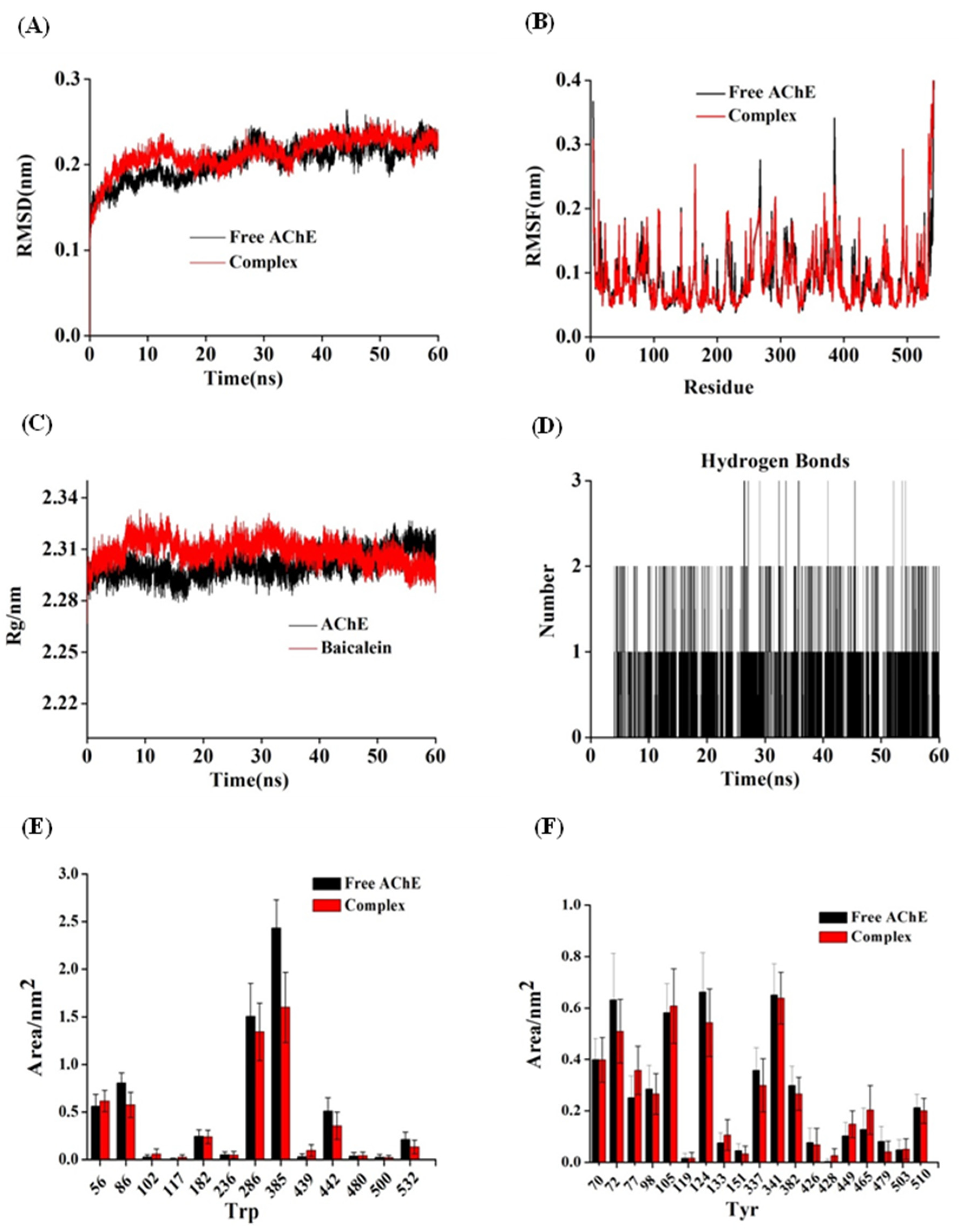
3.10. M.D. Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Scheltens, P.; Blennow, K.; Breteler, M.M.B.; de Strooper, B.; Frisoni, G.B.; Salloway, S.; Van der Flier, W.M. Alzheimer’s disease. Lancet 2016, 388, 505–517. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G. A Century of Alzheimer’s Disease. Science 2006, 314, 777–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauthier, S.; Rosa-Neto, P.; Morais, J.A.; Webster, C. World Alzheimer Report 2021: Journey through the Diagnosis of Dementia; Alzheimer’s Disease International: London, UK, 2021; Available online: https://www.alzint.org/resource/world-alzheimer-report-2021/ (accessed on 12 November 2021).
- Luo, W.; Wang, T.; Hong, C.; Yang, Y.C.; Chen, Y.; Cen, J.; Xie, S.Q.; Wang, C.J. Design, synthesis and evaluation of 4-dimethylamine flavonoid derivatives as potential multifunctional anti-Alzheimer agents. Eur. J. Med. Chem. 2016, 122, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.F.; Wang, M.J.; Shentu, J.Y.; Huang, C.H.; Bai, Y.J.; Pan, H.B.; Zhang, D.F.; Yuan, Z.J.; Zhang, H.; Xiao, X.; et al. Inhibition of acetylcholinesterase activity and beta-amyloid oligomer formation by 6-bromotryptamine A, a multi-target anti-Alzheimer’s molecule. Oncol. Lett. 2020, 19, 1593–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashford, J.W. Treatment of Alzheimer’s Disease: The Legacy of the Cholinergic Hypothesis, Neuroplasticity, and Future Directions. J. Alzheimer’s Dis. 2015, 47, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Allsop, D. Amyloici deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol. Sci. 1991, 12, 383–388. [Google Scholar] [CrossRef]
- Bartus, R.T.; Bernard Beer, R.L.; Lippa, A.S. The Cholinergic Hypothesis of Geriatric Memory Dysfunction. Science 1982, 217, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Mesulam, M.M.; Cuello, A.C.; Farlow, M.R.; Giacobini, E.; Grossberg, G.T.; Khachaturian, A.S.; Vergallo, A.; Cavedo, E.; Snyder, P.J.; et al. The cholinergic system in the pathophysiology and treatment of Alzheimer’s disease. Brain 2018, 141, 1917–1933. [Google Scholar] [CrossRef]
- Xu, Y.C.; Cheng, S.M.; Sussman, J.L.; Silman, I.; Jiang, H.L. Computational Studies on Acetylcholinesterases. Molecules 2017, 22, 1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sussman, J.L.; Silman, I. Computational studies on cholinesterases: Strengthening our understanding of the integration of structure, dynamics and function. Neuropharmacology 2020, 179, 108265. [Google Scholar] [CrossRef]
- Craig, L.A.; Hong, N.S.; McDonald, R.J. Revisiting the cholinergic hypothesis in the development of Alzheimer’s disease. Neurosci. Biobehav. Rev. 2011, 35, 1397–1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, B.U.; Tabrez, S. Management of Alzheimer’s disease—An insight of the enzymatic and other novel potential targets. Int. J. Biol. Macromol. 2017, 97, 700–709. [Google Scholar] [CrossRef]
- Woodruff-Pak, D.S.; Lander, C.; Geerts, H. Nicotinic Cholinergic Modulation Galantamine as a Prototype. CNS Drug Rev. 2002, 8, 405–426. [Google Scholar] [CrossRef]
- Sharma, K. Cholinesterase inhibitors as Alzheimer’s therapeutics. Mol. Med. Rep. 2019, 20, 1479–1487. [Google Scholar] [CrossRef] [Green Version]
- Birsan, R.I.; Wilde, P.; Waldron, K.W.; Rai, D.K. Anticholinesterase activities of different solvent extracts of Brewer’s Spent Grain. Foods 2021, 10, 930. [Google Scholar] [CrossRef]
- Jeon, S.G.; Song, E.J.; Lee, D.; Park, J.; Nam, Y.; Kim, J.I.; Moon, M. Traditional oriental medicines and Alzheimer’s disease. Aging Dis. 2019, 10, 307–328. [Google Scholar] [CrossRef] [Green Version]
- Khan, H.; Marya;Amin, S.; Kamal, M.A.; Patel, S. Flavonoids as acetylcholinesterase inhibitors: Current therapeutic standing and future prospects. Biomed. Pharmacother. 2018, 101, 860–870. [Google Scholar] [CrossRef]
- Beking, K.; Vieira, A. Flavonoid intake and disability-adjusted life years due to Alzheimer’s and related dementias: A population-based study involving twenty-three developed countries. Public Health Nutr. 2010, 13, 1403–1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.Y.H.; Sun, M.Y.Y.; Sommerville, N.; Ngan, M.P.; Ponomarev, E.D.; Lin, G.; Rudd, J.A. Soy flavonoids prevent cognitive deficits induced by intra-gastrointestinal administration of beta-amyloid. Food Chem. Toxicol. 2020, 141, 111396. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Tan, S.; Shan, Y.L.; Wang, Y.G.; Cai, W.; Huang, X.H.; Liao, X.Y.; Li, H.Y.; Zhang, L.; Zhang, B.J.; et al. Baicalein improves behavioral dysfunction induced by Alzheimer’s disease in rats. Neuropsychiatr. Dis. Treat. 2016, 12, 3145–3152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahu, B.D.; Mahesh Kumar, J.; Sistla, R. Baicalein, a bioflavonoid, prevents cisplatin-induced acute kidney injury by up-regulating antioxidant defenses and down-regulating the MAPKs and NF-κB Pathways. PLoS ONE 2015, 10, e0134139. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.H.; Yu, H.T.; Pu, X.P.; Du, G.H. Baicalein prevents 6-hydroxydopamine-induced mitochondrial dysfunction in SH-SY5Y cells via inhibition of mitochondrial oxidation and up-regulation of DJ-1 protein expression. Molecules 2013, 18, 14726–14738. [Google Scholar] [CrossRef] [Green Version]
- Patwardhan, R.S.; Sharma, D.; Thoh, M.; Checker, R.; Sandur, S.K. Baicalein exhibits anti-inflammatory effects via inhibition of NF-κB transactivation. Biochem. Pharmacol. 2016, 108, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Goc, A.; Niedzwiecki, A.; Rath, M. In vitro evaluation of antibacterial activity of phytochemicals and micronutrients against Borrelia burgdorferi and Borrelia garinii. J. Appl. Microbiol. 2015, 119, 1561–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Z.Q.; Zhu, S.M.; Han, X.; Wang, Z.; Wu, S.W.; Zheng, R.S. Baicalein inhibits hepatocellular carcinoma cells through suppressing the expression of CD24. Int. Immunopharmacol. 2015, 29, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cui, W.; Li, G.; Yuan, S.; Xu, D.; Hoi, M.P.; Lin, Z.; Dou, J.; Han, Y.; Lee, S.M. Baicalein protects against 6-OHDA-induced neurotoxicity through activation of Keap1/Nrf2/HO-1 and involving PKCalpha and PI3K/AKT signaling pathways. J. Agric. Food Chem. 2012, 60, 8171–8182. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, J.; Holscher, C. Therapeutic Potential of Baicalein in Alzheimer’s Disease and Parkinson’s Disease. CNS Drugs 2017, 31, 639–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Lin, G.; Zuo, Z. Pharmacological effects and pharmacokinetics properties of Radix Scutellariae and its bioactive flavones. Biopharm. Drug Dispos. 2011, 32, 427–445. [Google Scholar] [CrossRef]
- Gao, L.; Li, C.; Yang, R.Y.; Lian, W.W.; Fang, J.S.; Pang, X.C.; Qin, X.M.; Liu, A.L.; Du, G.H. Ameliorative effects of baicalein in MPTP-induced mouse model of Parkinson’s disease: A microarray study. Pharmacol. Biochem. Behav. 2015, 133, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.H.; Xu, L.J.; Liu, Z.Q.; Wei, B.; Yang, Y.J.; Xu, G.G.; Yin, X.P.; Wang, W. The flavonoid baicalein rescues synaptic plasticity and memory deficits in a mouse model of Alzheimer’s disease. Behav. Brain Res. 2016, 311, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.F.; Tang, J.F.; Bai, W.G.; Wang, Y.Y.; Zhang, Z.J. Ameliorative effects of baicalein on an amyloid-β induced Alzheimer’s disease rat model: A proteomics study. Curr. Alzheimer Res. 2014, 11, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Ji, Y.; Youn, K.; Lim, G.; Lee, J.; Kim, D.H.; Jun, M. Baicalein as a Potential Inhibitor against BACE1 and AChE: Mechanistic Comprehension through In Vitro and Computational Approaches. Nutrients 2019, 11, 2694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Yang, W.; Chen, X.; Xiao, J. Inhibition of flavonoids on acetylcholine esterase: Binding and structure-activity relationship. Food Funct. 2014, 5, 2582–2589. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L.; Courtney, K.D.; Andres, V.J.; Featherstone, J.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Ding, H.F.; Wu, X.Q.; Pan, J.H.; Hu, X.; Gong, D.M.; Zhang, G.W. New insights into the inhibition mechanism of betulinic acid on alpha-glucosidase. J. Agric. Food Chem. 2018, 66, 7065–7075. [Google Scholar] [CrossRef]
- Wang, Y.J.; Zhang, G.W.; Yan, J.K.; Gong, D.M. Inhibitory effect of morin on tyrosinase: Insights from spectroscopic and molecular docking studies. Food Chem. 2014, 163, 226–233. [Google Scholar] [CrossRef]
- Zeng, L.; Ding, H.F.; Hu, X.; Zhang, G.W.; Gong, D.M. Galangin inhibits α-glucosidase activity and formation of non-enzymatic glycation products. Food Chem. 2019, 271, 70–79. [Google Scholar] [CrossRef]
- Fan, M.H.; Zhang, G.W.; Hu, X.; Xu, X.M.; Gong, D.M. Quercetin as a tyrosinasm inhibitor: Inhibitory activity, conformational change and mechanism. Food Res. Int. 2017, 100, 226–233. [Google Scholar] [CrossRef]
- Ni, M.T.; Hu, X.; Gong, D.M.; Zhang, G.W. Inhibitory mechanism of vitexin on α-glucosidase and its synergy with acarbose. Food Hydrocoll. 2020, 105, 105824. [Google Scholar] [CrossRef]
- Cheung, J.; Rudolph, M.J.; Burshteyn, F.; Cassidy, M.S.; Gary, E.N.; Love, J.; Franklin, M.C.; Height, J.J. Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J. Med. Chem. 2012, 55, 10282–10286. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandar, N.B.; Efremenko, I.; Silman, I.; Martin, J.M.L.; Sussman, J.L. Molecular dynamics simulations of the interaction of Mouse and Torpedo acetylcholinesterase with covalent inhibitors explain their differential reactivity: Implications for drug design. Chem. Biol. Interact. 2019, 310, 108715. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Hu, X.; Zhang, Y.; Pan, J.H.; Gong, D.M.; Zhang, G.W. Inhibitory mechanism of epicatechin gallate on tyrosinase: Inhibitory interaction, conformational change and computational simulation. Food Funct. 2020, 11, 4892–4902. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.B.; Cao, H.; Wang, Y.F.; Zhao, J.Y.; Wei, X.L. Glycosylation of dietary flavonoids decreases the affinities for plasma protein. J. Agric. Food Chem. 2009, 57, 6642–6648. [Google Scholar] [CrossRef]
- Cao, H.; Liu, X.J.; Ulrih, N.P.; Sengupta, P.K.; Xiao, J.B. Plasma protein binding of dietary polyphenols to human serum albumin: A high performance affinity chromatography approach. Food Chem. 2019, 270, 257–263. [Google Scholar] [CrossRef]
- Katalinic, M.; Rusak, G.; Domacinovic Barovic, J.; Sinko, G.; Jelic, D.; Antolovic, R.; Kovarik, Z. Structural aspects of flavonoids as inhibitors of human butyrylcholinesterase. Eur. J. Med. Chem. 2010, 45, 186–192. [Google Scholar] [CrossRef]
- Han, L.; Fang, C.; Zhu, R.; Peng, Q.; Li, D.; Wang, M. Inhibitory effect of phloretin on α-glucosidase: Kinetics, interaction mechanism and molecular docking. Int. J. Biol. Macromol. 2017, 95, 520–527. [Google Scholar] [CrossRef]
- Lin, S.Y.; Zhang, G.W.; Liao, Y.J.; Pan, J.H. Inhibition of chrysin on xanthine oxidase activity and its inhibition mechanism. Int. J. Biol. Macromol. 2015, 81, 274–282. [Google Scholar] [CrossRef]
- Jin, Q.X.; Yin, S.J.; Wang, W.; Wang, Z.J.; Yang, J.M.; Qian, G.Y.; Si, Y.X.; Park, Y.D. The effect of Zn2+ on Euphausia superba arginine kinase: Unfolding and aggregation studies. Process Biochem. 2014, 49, 821–829. [Google Scholar] [CrossRef]
- Yan, J.K.; Zhang, G.W.; Pan, J.H.; Wang, Y.J. α-Glucosidase inhibition by luteolin: Kinetics, interaction and molecular docking. Int. J. Biol. Macromol. 2014, 64, 213–223. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, P.; Ying, J.; Dong, Z.; Chen, X.D. Mechanistic study on inhibition of porcine pancreatic α-amylase using the flavonoids from dandelion. Food Chem. 2021, 344, 128610. [Google Scholar] [CrossRef]
- Zeng, L.; Zhang, G.W.; Lin, S.Y.; Gong, D.M. Inhibitory mechanism of apigenin on α-glucosidase and synergy analysis of flavonoids. J. Agric. Food Chem. 2016, 64, 6939–6949. [Google Scholar] [CrossRef]
- Danesh, N.; Navaee Sedighi, Z.; Beigoli, S.; Sharifi-Rad, A.; Saberi, M.R.; Chamani, J. Determining the binding site and binding affinity of estradiol to human serum albumin and holo-transferrin: Fluorescence spectroscopic, isothermal titration calorimetry and molecular modeling approaches. J. Biomol. Struct. Dyn. 2018, 36, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Chai, W.M.; Lin, M.Z.; Wang, Y.X.; Xu, K.L.; Huang, W.Y.; Pan, D.D.; Zou, Z.R.; Peng, Y.Y. Inhibition of tyrosinase by cherimoya pericarp proanthocyanidins: Structural characterization, inhibitory activity and mechanism. Food Res. Int. 2017, 100, 731–739. [Google Scholar] [CrossRef] [PubMed]
- He, X.M.; Carter, D.C. Atomic structure and chemistry of human serum albumin. Nature 1992, 358, 209–214. [Google Scholar] [CrossRef] [Green Version]
- Dai, T.T.; Chen, J.; McClements, D.J.; Li, T.; Liu, C.M. Investigation the interaction between procyanidin dimer and α-glucosidase: Spectroscopic analyses and molecular docking simulation. Int. J. Biol. Macromol. 2019, 130, 315–322. [Google Scholar] [CrossRef]
- Wu, X.Q.; Ding, H.F.; Hu, X.; Pan, J.H.; Liao, Y.J.; Gong, D.M.; Zhang, G.W. Exploring inhibitory mechanism of gallocatechin gallate on a-amylase and a-glucosidase relevant to postprandial hyperglycemia. J. Funct. Foods 2018, 48, 200–209. [Google Scholar] [CrossRef]
- Bertucci, C.; Pistolozzi, M.; De Simone, A. Circular dichroism in drug discovery and development: An abridged review. Anal. Bioanal. Chem. 2010, 398, 155–166. [Google Scholar] [CrossRef]
- Manavalan, P.; Taylor, P.; Johnson, W.C., Jr. Circular dichroism studies of acetylcholinesterase conformation. Comparison of the 11 S and 5.6 S species and the differences induced by inhibitory ligands. Biochim. Biophys. Acta 1985, 829, 365–370. [Google Scholar] [CrossRef]
- Golicnik, M.; Fournier, D.; Stojan, J. Interaction of drosophila acetylcholinesterases with d-Tubocurarine: An explanation of the activation by an inhibitor. Biochemistry 2001, 40, 1214–1219. [Google Scholar] [CrossRef]
- Zhu, M.; Pan, J.H.; Hu, X.; Zhang, G.W. Epicatechin gallate as xanthine oxidase inhibitor: Inhibitory kinetics, binding characteristics, synergistic inhibition, and action mechanism. Foods 2021, 10, 2191. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Ni, M.T.; Zhang, Y.; Zhang, G.W.; Pan, J.H.; Gong, D.M. Comparing the inhibitory abilities of epigallocatechin-3-gallate and gallocatechin gallate against tyrosinase and their combined effects with kojic acid. Food Chem. 2021, 349, 129172. [Google Scholar] [CrossRef] [PubMed]
- Sahihi, M.; Ghayeb, Y. An investigation of molecular dynamics simulation and molecular docking: Interaction of citrus flavonoids and bovine beta-lactoglobulin in focus. Comput. Biol. Med. 2014, 51, 44–50. [Google Scholar] [CrossRef] [PubMed]

| Baicalein (µM) | Inactivation Rate Constant a (×10−4 s−1) | Transition Free Energy Change b (kJ mol−1 s−1) |
|---|---|---|
| k | ||
| 2 | 1.48 ± 0.03 | 21.85 |
| 4 | 2.42 ± 0.05 | 20.63 |
| 8 | 4.01 ± 0.02 | 19.38 |
| 16 | 5.18 ± 0.08 | 18.74 |
| T (°C) | KSV (×104 L mol−1) | Ra | Ka (×104 L mol−1) | Rb | n | ∆H° (kJ mol−1) | ∆G° (kJ mol−1) | ∆S° (J mol−1 K−1) |
|---|---|---|---|---|---|---|---|---|
| 25 | 6.45 ± 0.02 | 0.9989 | 6.66 ± 0.05 | 0.9982 | 0.88 ± 0.01 | −17.01 ± 0.02 | −22.71 ± 0.06 | 19.15 ± 0.09 |
| 31 | 5.78± 0.03 | 0.9972 | 5.84 ± 0.01 | 0.9979 | 0.93 ± 0.03 | −22.83 ± 0.03 | ||
| 37 | 5.22± 0.07 | 0.9946 | 5.11 ± 0.04 | 0.9967 | 1.13 ± 0.02 | −22.94 ± 0.08 |
| Molar Ratio (Baicalein):(AChE) | α-Helix (%) | β-Sheet (%) | β-Turn (%) | Random Coil (%) |
|---|---|---|---|---|
| 0:1 | 34.35 ± 0.74 | 18.62 ± 0.06 | 19.18 ± 0.28 | 28.02 ± 0.65 |
| 2:1 | 38.82 ± 0.08 | 14.57 ± 0.13 | 21.42 ± 0.57 | 25.36 ± 0.29 |
| 6:1 | 42.53 ± 0.35 | 7.73 ± 0.03 | 28.15 ± 0.89 | 21.71 ± 0.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, Y.; Hu, X.; Pan, J.; Zhang, G. Inhibitory Mechanism of Baicalein on Acetylcholinesterase: Inhibitory Interaction, Conformational Change, and Computational Simulation. Foods 2022, 11, 168. https://doi.org/10.3390/foods11020168
Liao Y, Hu X, Pan J, Zhang G. Inhibitory Mechanism of Baicalein on Acetylcholinesterase: Inhibitory Interaction, Conformational Change, and Computational Simulation. Foods. 2022; 11(2):168. https://doi.org/10.3390/foods11020168
Chicago/Turabian StyleLiao, Yijing, Xing Hu, Junhui Pan, and Guowen Zhang. 2022. "Inhibitory Mechanism of Baicalein on Acetylcholinesterase: Inhibitory Interaction, Conformational Change, and Computational Simulation" Foods 11, no. 2: 168. https://doi.org/10.3390/foods11020168
APA StyleLiao, Y., Hu, X., Pan, J., & Zhang, G. (2022). Inhibitory Mechanism of Baicalein on Acetylcholinesterase: Inhibitory Interaction, Conformational Change, and Computational Simulation. Foods, 11(2), 168. https://doi.org/10.3390/foods11020168








