Dissecting of the AI-2/LuxS Mediated Growth Characteristics and Bacteriostatic Ability of Lactiplantibacillus plantarum SS-128 by Integration of Transcriptomics and Metabolomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of luxS-Mutant of SS-128
2.2. AI-2 Assay
2.3. Bacterial Growth Assay
2.4. Flow Cytometry Assay
2.5. Determination of Bacteriostatic Ability
2.6. Transcriptome Analysis
2.7. Non-Targeted Metabolomic Analysis
2.7.1. Sample Preparation
2.7.2. Gas Chromatography/Mass Spectrometry (GC/MS) Analysis
2.7.3. Ultrahigh Performance Liquid Chromatography/Mass Spectrometry (UPLC/MS) Analysis
2.7.4. Data Preprocessing and Analysis
2.8. Statistical Analysis
3. Results
3.1. Confirmation of luxS Gene Knockout
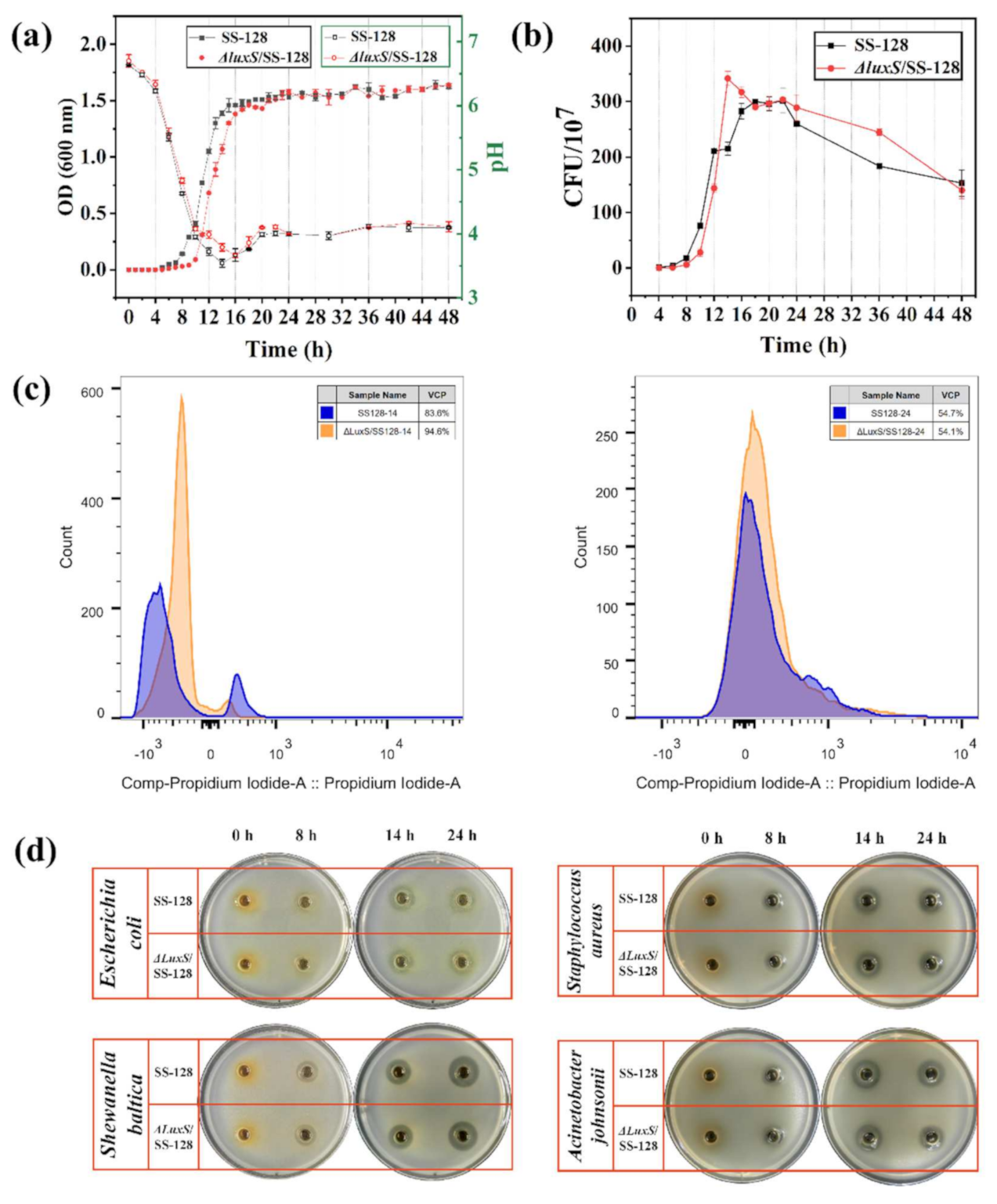
3.2. Growth Characteristics of L. plantarum SS-128 and ΔluxS/SS-128
3.3. In Vitro Bacteriostatic Effect of L. plantarum SS-128 and ΔluxS/SS-128
3.4. The Organic Acids Production of L. plantarum SS-128 and ΔluxS/SS-128
3.5. Transcriptomics Analyses between L. plantarum SS-128 and the ΔluxS/SS-128
3.6. Metabolomics Analyses between L. plantarum SS-128 and the ΔluxS/SS-128
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hatti-Kaul, R.; Chen, L.; Dishisha, T.; Enshasy, H.E. Lactic acid bacteria: From starter cultures to producers of chemicals. FEMS Microbiol. Lett. 2018, 365, fny213. [Google Scholar] [CrossRef] [PubMed]
- Barcenilla, C.; Ducic, M.; López, M.; Prieto, M.; Álvarez-Ordóñez, A. Application of lactic acid bacteria for the biopreservation of meat products: A systematic review. Meat Sci. 2022, 183, 108661. [Google Scholar] [CrossRef] [PubMed]
- Crowley, S.; Mahony, J.; van Sinderen, D. Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives. Trends Food Sci. Technol. 2013, 33, 93–109. [Google Scholar] [CrossRef]
- Nasrollahzadeh, A.; Mokhtari, S.; Khomeiri, M.; Saris, P.E. Antifungal Preservation of Food by Lactic Acid Bacteria. Foods 2022, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Michon, C.; Langella, P.; Eijsink, V.G.; Mathiesen, G.; Chatel, J.M. Display of recombinant proteins at the surface of lactic acid bacteria: Strategies and applications. Microb. Cell Fact. 2016, 15, 70. [Google Scholar] [CrossRef]
- Gao, C.; Ma, C.; Xu, P. Biotechnological routes based on lactic acid production from biomass. Biotechnol. Adv. 2011, 29, 930–939. [Google Scholar] [CrossRef]
- Gálvez, A.; Abriouel, H.; Benomar, N.; Lucas, R. Microbial antagonists to food-borne pathogens and biocontrol. Curr. Opin. Biotechnol. 2010, 21, 142–148. [Google Scholar] [CrossRef]
- Kos, B.; Šušković, J.; Beganović, J.; Gjuračić, K.; Frece, J.; Iannaccone, C.; Canganella, F. Characterization of the three selected probiotic strains for the application in food industry. World J. Microbiol. Biotechnol. 2008, 24, 699–707. [Google Scholar] [CrossRef]
- Lee, S.-J.; Jeon, H.-S.; Yoo, J.-Y.; Kim, J.-H. Some Important Metabolites Produced by Lactic Acid Bacteria Originated from Kimchi. Foods 2021, 10, 2148. [Google Scholar] [CrossRef]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef]
- Schauder, S.; Shokat, K.; Surette, M.G.; Bassler, B.L. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 2001, 41, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, H.; Xie, Y.; Guo, Y.; Cheng, Y.; Yao, W. Quorum sensing inhibitory effect of hexanal on Autoinducer-2 (AI-2) and corresponding impacts on biofilm formation and enzyme activity in Erwinia carotovora and Pseudomonas fluorescens isolated from vegetables. J. Food Processing Preserv. 2022, 46, e16293. [Google Scholar] [CrossRef]
- Pereira, C.S.; Thompson, J.A.; Xavier, K.B. AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 2013, 37, 156–181. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, B.; Tian, J.; Wu, R.; He, Y. The response of LuxS/AI-2 quorum sensing in Lactobacillus fermentum 2-1 to changes in environmental growth conditions. Ann. Microbiol. 2018, 68, 287–294. [Google Scholar] [CrossRef]
- Gu, Y.; Wu, J.; Tian, J.; Li, L.; Zhang, B.; Zhang, Y.; He, Y. Effects of Exogenous Synthetic Autoinducer-2 on Physiological Behaviors and Proteome of Lactic Acid Bacteria. ACS Omega 2020, 5, 1326–1335. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Tian, J.; Zhang, Y.; Wu, R.; Li, L.; Zhang, B.; He, Y. Dissecting signal molecule AI-2 mediated biofilm formation and environmental tolerance in Lactobacillus plantarum. J. Biosci. Bioeng. 2021, 131, 153–160. [Google Scholar] [CrossRef]
- Yeo, S.; Park, H.; Ji, Y.; Park, S.; Yang, J.; Lee, J.; Mathara, J.M.; Shin, H.; Holzapfel, W. Influence of gastrointestinal stress on autoinducer-2 activity of two Lactobacillus species. FEMS Microbiol. Ecol. 2015, 91, fiv065. [Google Scholar] [CrossRef]
- Man, L.-L.; Meng, X.-C.; Zhao, R.-H.; Xiang, D.-J. The role of plNC8HK-plnD genes in bacteriocin production in Lactobacillus plantarum KLDS1.0391. Int. Dairy J. 2014, 34, 267–274. [Google Scholar] [CrossRef]
- Tannock, G.W.; Ghazally, S.; Walter, J.; Loach, D.; Brooks, H.; Cook, G.; Surette, M.; Simmers, C.; Bremer, P.; Dal Bello, F.; et al. Ecological behavior of Lactobacillus reuteri 100-23 is affected by mutation of the luxS gene. Appl. Environ. Microbiol. 2005, 71, 8419–8425. [Google Scholar] [CrossRef]
- Lebeer, S.; De Keersmaecker, S.C.; Verhoeven, T.L.; Fadda, A.A.; Marchal, K.; Vanderleyden, J. Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation. J. Bacteriol. 2007, 189, 860–871. [Google Scholar] [CrossRef]
- Jia, F.F.; Pang, X.H.; Zhu, D.Q.; Zhu, Z.T.; Sun, S.R.; Meng, X.C. Role of the luxS gene in bacteriocin biosynthesis by Lactobacillus plantarum KLDS1.0391: A proteomic analysis. Sci. Rep. 2017, 7, 13871. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, Y.; Guo, X.; Zhang, L.; Zhang, W.; Man, C.; Jiang, Y. Characterization and transcriptomic basis of biofilm formation by Lactobacillus plantarum J26 isolated from traditional fermented dairy products. LWT 2020, 125, 109333. [Google Scholar] [CrossRef]
- Li, J.; Yang, X.; Shi, G.; Chang, J.; Liu, Z.; Zeng, M. Cooperation of lactic acid bacteria regulated by the AI-2/LuxS system involve in the biopreservation of refrigerated shrimp. Food Res. Int. 2019, 120, 679–687. [Google Scholar] [CrossRef]
- Bassler, B.L.; Greenberg, E.P.; Stevens, A.M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 1997, 179, 4043–4045. [Google Scholar] [CrossRef]
- Léonard, L.; Beji, O.; Arnould, C.; Noirot, E.; Bonnotte, A.; Gharsallaoui, A.; Degraeve, P.; Lherminier, J.; Saurel, R.; Oulahal, N. Preservation of viability and anti-Listeria activity of lactic acid bacteria, Lactococcus lactis and Lactobacillus paracasei, entrapped in gelling matrices of alginate or alginate/caseinate. Food Control 2015, 47, 7–19. [Google Scholar] [CrossRef]
- Bensch, G.; Rüger, M.; Wassermann, M.; Weinholz, S.; Reichl, U.; Cordes, C. Flow cytometric viability assessment of lactic acid bacteria starter cultures produced by fluidized bed drying. Appl. Microbiol. Biotechnol. 2014, 98, 4897–4909. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, S.; Shi, Y.; Shen, F.; Wang, H. A new high phenyl lactic acid-yielding Lactobacillus plantarum IMAU10124 and a comparative analysis of lactate dehydrogenase gene. FEMS Microbiol. Lett. 2014, 356, 89–96. [Google Scholar] [CrossRef]
- Li, H.; Xie, X.; Li, Y.; Chen, M.; Xue, L.; Wang, J.; Zhang, J.; Wu, S.; Ye, Q.; Zhang, S.; et al. Pediococcus pentosaceus IM96 Exerts Protective Effects against Enterohemorrhagic Escherichia coli O157:H7 Infection. In Vivo Foods 2021, 10, 2945. [Google Scholar] [CrossRef]
- Kaur, A.; Capalash, N.; Sharma, P. Expression of Meiothermus ruber luxS in E. coli alters the antibiotic susceptibility and biofilm formation. Appl. Microbiol. Biotechnol. 2020, 104, 4457–4469. [Google Scholar] [CrossRef]
- Monahan, L.G.; Harry, E.J. You Are What You Eat: Metabolic Control of Bacterial Division. Trends Microbiol. 2016, 24, 181–189. [Google Scholar] [CrossRef]
- Rang, U.; Peng, A.; Poon, A.; Chao, L. Ageing in Escherichia coli requires damage by an extrinsic agent. Microbiology 2012, 158, 1553–1559. [Google Scholar] [CrossRef] [PubMed]
- Nakaoka, H.; Wakamoto, Y. Aging, mortality, and the fast growth trade-off of Schizosaccharomyces pombe. PLoS Biol. 2017, 15, e2001109. [Google Scholar] [CrossRef]
- Rathod, N.B.; Phadke, G.G.; Tabanelli, G.; Mane, A.; Ranveer, R.C.; Pagarkar, A.; Ozogul, F. Recent advances in bio-preservatives impacts of lactic acid bacteria and their metabolites on aquatic food products. Food Biosci. 2021, 44, 101440. [Google Scholar] [CrossRef]
- El-Saber Batiha, G.; Hussein, D.E.; Algammal, A.M.; George, T.T.; Jeandet, P.; Al-Snafi, A.E.; Tiwari, A.; Pagnossa, J.P.; Lima, C.M.; Thorat, N.D.; et al. Application of natural antimicrobials in food preservation: Recent views. Food Control 2021, 126, 108066. [Google Scholar] [CrossRef]
- Shekarforoush, S.S.; Basiri, S.; Ebrahimnejad, H.; Hosseinzadeh, S. Effect of chitosan on spoilage bacteria, Escherichia coli and Listeria monocytogenes in cured chicken meat. Int. J. Biol. Macromol. 2015, 76, 303–309. [Google Scholar] [CrossRef]
- Hong Tran, D.; Tran, T.; Pham, N.; Phung, H. Direct multiplex recombinase polymerase amplification for rapid detection of Staphylococcus aureus and Pseudomonas aeruginosa in food. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ge, Y.; Zhu, J.; Ye, X.; Yang, Y. Spoilage potential characterization of Shewanella and Pseudomonas isolated from spoiled large yellow croaker (Pseudosciaena crocea). Lett. Appl. Microbiol. 2017, 64, 86–93. [Google Scholar] [CrossRef]
- Wang, X.-Y.; Xie, J. Quorum Sensing System-Regulated Proteins Affect the Spoilage Potential of Co-cultured Acinetobacter johnsonii and Pseudomonas fluorescens From Spoiled Bigeye Tuna (Thunnus obesus) as Determined by Proteomic Analysis. Front. Microbiol. 2020, 11, 940. [Google Scholar] [CrossRef]
- Flickinger, S.T.; Copeland, M.F.; Downes, E.M.; Braasch, A.T.; Tuson, H.H.; Eun, Y.-J.; Weibel, D.B. Quorum sensing between Pseudomonas aeruginosa biofilms accelerates cell growth. J. Am. Chem. Soc. 2011, 133, 5966–5975. [Google Scholar] [CrossRef]
- Jie, J.; Yu, H.; Han, Y.; Liu, Z.; Zeng, M. Acyl-homoserine-lactones receptor LuxR of Shewanella baltica involved in the development of microbiota and spoilage of refrigerated shrimp. J. Food Sci. Technol. 2018, 55, 2795–2800. [Google Scholar] [CrossRef]
- Li, X.; Jiang, B.; Pan, B.; Mu, W.; Zhang, T. Purification and partial characterization of Lactobacillus species SK007 lactate dehydrogenase (LDH) catalyzing phenylpyruvic acid (PPA) conversion into phenyllactic acid (PLA). J. Agric. Food Chem. 2008, 56, 2392–2399. [Google Scholar] [CrossRef]
- Lin, M.; Zhou, G.-H.; Wang, Z.-G.; Yun, B. Functional analysis of AI-2/LuxS from bacteria in Chinese fermented meat after high nitrate concentration shock. Eur. Food Res. Technol. 2015, 240, 119–127. [Google Scholar] [CrossRef]
- Moslehi-Jenabian, S.; Vogensen, F.K.; Jespersen, L. The quorum sensing luxS gene is induced in Lactobacillus acidophilus NCFM in response to Listeria monocytogenes. Int. J. Food Microbiol. 2011, 149, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Fontecave, M.; Atta, M.; Mulliez, E. S-adenosylmethionine: Nothing goes to waste. Trends Biochem. Sci. 2004, 29, 243–249. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Z.; Cai, H.; Zhou, C. Progress in the microbial production of S-adenosyl-L-methionine. World J. Microbiol. Biotechnol. 2016, 32, 153. [Google Scholar] [CrossRef] [PubMed]
- Kilstrup, M.; Hammer, K.; Ruhdal Jensen, P.; Martinussen, J. Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol. Rev. 2005, 29, 555–590. [Google Scholar] [CrossRef] [PubMed]
- Vilain, S.; Pretorius, J.M.; Theron, J.; Brözel, V.S. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl. Environ. Microb 2009, 75, 2861–2868. [Google Scholar] [CrossRef] [PubMed]
- Duwat, P.; Ehrlich, S.D.; Gruss, A. Effects of metabolic flux on stress response pathways in Lactococcus lactis. Mol. Microbiol. 1999, 31, 845–858. [Google Scholar] [CrossRef]
- Rallu, F.; Gruss, A.; Ehrlich, S.D.; Maguin, E. Acid- and multistress-resistant mutants of Lactococcus lactis: Identification of intracellular stress signals. Mol. Microbiol. 2000, 35, 517–528. [Google Scholar] [CrossRef]
- Nicoloff, H.; Hubert, J.-C.; Bringel, F. In Lactobacillus plantarum, Carbamoyl Phosphate Is Synthesized by Two Carbamoyl-Phosphate Synthetases (CPS): Carbon Dioxide Differentiates the Arginine-Repressed from the Pyrimidine-Regulated CPS. J. Bacteriol. 2000, 182, 3416–3422. [Google Scholar] [CrossRef]
- Sperandio, B.; Polard, P.; Ehrlich, D.S.; Renault, P.; Guédon, E. Sulfur amino acid metabolism and its control in Lactococcus lactis IL1403. J. Bacteriol. 2005, 187, 3762–3778. [Google Scholar] [CrossRef] [PubMed]
- Owen, O.E.; Kalhan, S.C.; Hanson, R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002, 277, 30409–30412. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, A.R.; Alkema, W.; Starrenburg, M.J.; Hugenholtz, J.; van Hijum, S.A.; Bron, P.A. Fermentation-induced variation in heat and oxidative stress phenotypes of Lactococcus lactis MG1363 reveals transcriptome signatures for robustness. Microb Cell Fact. 2014, 13, 148. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kornberg, H.L. The role and control of the glyoxylate cycle in Escherichia coli. Biochem. J. 1966, 99, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Petersen, S.; de Graaf, A.A.; Eggeling, L.; Möllney, M.; Wiechert, W.; Sahm, H. In Vivo Quantification of Parallel and Bidirectional Fluxes in the Anaplerosis of Corynebacterium glutamicum. J. Biol. Chem. 2000, 275, 35932–35941. [Google Scholar] [CrossRef] [PubMed]
- Reis, J.A.; Paula, A.T.; Casarotti, S.N.; Penna, A.L.B. Lactic Acid Bacteria Antimicrobial Compounds: Characteristics and Applications. Food Eng. Rev. 2012, 4, 124–140. [Google Scholar] [CrossRef]
- Yang, X.; Li, J.; Shi, G.; Zeng, M.; Liu, Z. Improving 3-phenyllactic acid production of Lactobacillus plantarum AB-1 by enhancing its quorum-sensing capacity. J. Food Sci. Technol. 2019, 56, 2605–2610. [Google Scholar] [CrossRef]
- Feldman-Salit, A.; Hering, S.; Messiha, H.L.; Veith, N.; Cojocaru, V.; Sieg, A.; Westerhoff, H.V.; Kreikemeyer, B.; Wade, R.C.; Fiedler, T. Regulation of the activity of lactate dehydrogenases from four lactic acid bacteria. J. Biol. Chem. 2013, 288, 21295–21306. [Google Scholar] [CrossRef]
- Rajanikar, R.V.; Nataraj, B.H.; Naithani, H.; Ali, S.A.; Panjagari, N.R.; Behare, P.V. Phenyllactic acid: A green compound for food biopreservation. Food Control 2021, 128, 108184. [Google Scholar] [CrossRef]
- Jia, J.; Mu, W.; Zhang, T.; Jiang, B. Bioconversion of Phenylpyruvate to Phenyllactate: Gene Cloning, Expression, and Enzymatic Characterization of d-and l 1-Lactate Dehydrogenases from Lactobacillus plantarum SK002. Appl. Biochem. Biotechnol. 2010, 162, 242–251. [Google Scholar] [CrossRef]
- Stoll, R.; Goebel, W. The major PEP-phosphotransferase systems (PTSs) for glucose, mannose and cellobiose of Listeria monocytogenes, and their significance for extra- and intracellular growth. Microbiology 2010, 156, 1069–1083. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Thompson, C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef] [PubMed]





| Gene Name | Oligonucleotide Sequence (5′-3′) | |
|---|---|---|
| Up | Forward | TATCCC ACTACCTGAA ACTCG |
| Reverse | GCACCACCATTACTTTTTATATTGTAGCACATTGCCCGTTA | |
| Down | Forward | TAACGGGCAATGTGCATACAATATAAAAAGTAATGGTGGTGC |
| Reverse | GCTGGTGCTTCGTAAACTTCC | |
| 16S rRNA | Forward | CGTAGGTGGCAAGCGTTGTCC |
| Reverse | CGCCTTCGCCACTGGTGTTC | |
| LuxS | Forward | CGGATGGATGGCGTGATTGACTG |
| Reverse | CTTAGCAACTTCAACGGTGTCATGTTC | |
| UD | Forward | GTTCTGCACGGACGCTATCT |
| Reverse | ATTAACTTGCGTTGGTAGGC | |
| Locus Tag | Entry | Gene Name (ID) | Definition | Fold Change | p-Value |
|---|---|---|---|---|---|
| PTS | K02793 | manXa | mannose PTS system EIIA component | 1.56 ↓ | 1.12 × 104 |
| K02768 | fruB | fructose PTS system EIIA component | 1.68 ↓ | 2.14 × 106 | |
| K02810 | scrA, ptsS | sucrose PTS system EIIBCA component | 1.54 ↓ | 8.15 × 103 | |
| K02761 | celB, chbC | cellobiose PTS system EIIC component | 3.36 ↓ * | 2.21 × 104 | |
| K02760 | celA, chbB | cellobiose PTS system EIIB component | 1.92 ↓ | 6.99 × 104 | |
| K02759 | celC, chbA | cellobiose PTS system EIIA component | 3.19 ↓* | 4.32 × 104 | |
| K02773 | gatA, sgcA | galactitol PTS system EIIA component | 1.88 ↓ | 2.76 × 104 | |
| K02774 | gatB, sgcB | galactitol PTS system EIIB component | 1.71 ↓ | 1.46 × 102 | |
| K02755 | bglF | beta-glucoside PTS system EIIA component | 5.93 ↓ * | 3.28 × 109 | |
| K02798 | cmtB | mannitol PTS system EIIA component | 1.59 ↓ | 1.49 × 105 | |
| LP_RS14755 | PTS system EIIC component | 1.74 ↓ | 1.79 × 102 | ||
| LP_RS12340 | PTS system EIIA component | 1.67 ↓ | 3.06 × 102 | ||
| LP_RS12650 | PTS system EIIB component | 1.52 ↓ | 4.89 × 102 | ||
| LP_RS12655 | PTS system EIIC component | 2.33 ↓ * | 9.25× 1010 | ||
| LP_RS13505 | beta-glucoside PTS system EIIBCA component | 1.54 ↓ | 8.15× 103 | ||
| LP_RS14845 | PTS system EIIB component | 1.71 ↓ | 1.46 × 102 | ||
| LP_RS14850 | PTS system EIIA component | 1.88 ↓ | 2.76 × 104 | ||
| Pyruvate metabolism | K00927 | PGK, pgk | phosphoglycerate kinase | 1.52 ↑ | 1.85 × 105 |
| K00016 | ldh | L-lactate dehydrogenase | 2.58 ↓ * | 1.03 × 102 | |
| K01610 | pckA, PEPCK | phosphoenolpyruvate carboxykinase (ATP) | 1.51 ↑ | 3.95 × 102 | |
| K00027 | ME, maeA | malate dehydrogenase | 4.87 ↑ * | 5.68 × 103 | |
| K01676 | fumA, fumB | fumarate hydratase, class I | 3.39 ↑ * | 1.10 × 104 | |
| K00244 | frdA | fumarate reductase flavoprotein subunit | 4.08 ↑ * | 3.14 × 102 | |
| K01744 | aspA | aspartate ammonia-lyase | 1.70 ↑ | 1.99 × 107 | |
| K01939 | purA, ADSS | adenylosuccinate synthase | 1.55 ↑ | 2.25 × 102 | |
| K01512 | acyP | acylphosphatase | 1.70 ↑ | 2.95 × 105 | |
| Methionine cycle | K07173 | luxS | S-ribosylhomocysteine lyase | 2.53 × 104 | |
| K01738 | cysK | cysteine synthase | 2.95 ↓ | 5.85 × 1010 | |
| K01999 | livK | branched-chain amino acid transport system substrate-binding protein | 2.01 ↓ * | 2.75 × 102 |
| Metabolites | KEGG | KEGG Annotation | Dataclass | Formula | VIP | p-Value | FC |
|---|---|---|---|---|---|---|---|
| 2′-Deoxyguanosine 5′-monophosphate | C00362 | Purine metabolism | LC | C10H14N5O7P | 1.12 | 3.46 × 108 | 1.53 ↑ |
| Deoxyguanosine | C00330 | Purine metabolism | LC | C10H13N5O4 | 1.26 | 2.38 × 106 | 1.84 ↑ |
| Guanine | C00242 | Purine metabolism | GC | C5H5N5O | 1.38 | 6.32 × 104 | 2.05 ↑ * |
| Hypoxanthine | C00262 | Purine metabolism | GC | C5H4N4O2 | 2.27 | 4.52 × 105 | 5.61 ↑ * |
| Xanthine | C00385 | Purine metabolism | LC | C5H4N4O2 | 1.14 | 1.08 × 107 | 2.21 ↑ * |
| Cytosine | C00380 | Pyrimidine metabolism | GC | C4H5N3O | 1.43 | 5.73 × 104 | 2.15 ↑ * |
| dCMP | C00239 | Pyrimidine metabolism | LC | C9H14N3O7P | 1.62 | 1.58 × 104 | 1.62 ↑ |
| dTMP | C00364 | Pyrimidine metabolism | LC | C10H15N2O8P | 5.82 | 1.20 × 107 | 1.73 ↑ |
| Pseudouridine 5′-phosphate | C01168 | Pyrimidine metabolism | LC | C9H13N2O9P | 4.18 | 6.41 × 105 | 1.15 ↑ |
| Uracil | C00106 | Pyrimidine metabolism | GC | C4H4N2O2 | 1.67 | 3.17 × 105 | 2.59 ↑ * |
| Uridine | C00299 | Pyrimidine metabolism | GC | C9H12N2O6 | 1.29 | 3.43 × 104 | 1.86 ↑ |
| Uridine 5′-monophosphate | C00105 | Pyrimidine metabolism | LC | C9H13N2O9P | 2.20 | 5.38 × 104 | 1.42 ↑ |
| Orotidine | C01103 | Pyrimidine metabolism | LC | C10H12N2O8 | 1.42 | 7.33 × 107 | 4.61 ↓ * |
| L-glutamine | C00064 | Purine metabolism pyrimidine metabolism | GC | C5H10N2O3 | 3.13 | 3.58 × 107 | 22.12 ↓ * |
| Flavin Mononucleotide | C00061 | Oxidative phosphorylation Riboflavin metabolism | LC | C17H21N4O9P | 1.04 | 4.29 × 103 | 1.66 ↑ |
| Riboflavin | C00255 | ABC transporters Riboflavin metabolism | LC | C17H20N4O6 | 1.23 | 3.40 × 103 | 2.22 ↑ * |
| Flavin adenine dinucleotide | C00016 | Riboflavin metabolism | LC | C27H33N9O15P2 | 2.96 | 2.77 × 1010 | 1.64 ↑ |
| D-Glycerate 2-phosphate | C00631 | Glycolysis/Gluconeogenesis | LC | C3H7O7P | 1.81 | 2.85 × 107 | 1.84 ↑ |
| D-Glycerate 3-phosphate | C00197 | Glycolysis/Gluconeogenesis | LC | C3H7O7P | 1.10 | 9.00 × 108 | 1.79 ↑ |
| Phosphoenolpyruvic acid | C00074 | Glycolysis/Gluconeogenesis Citrate cycle (TCA cycle) | LC | C3H5O6P | 1.86 | 5.37 × 108 | 1.91 ↑ |
| Isocitric acid | C00311 | Citrate cycle (TCA cycle) | GC | C6H8O7 | 1.22 | 2.09 × 102 | 4.97 ↑ * |
| Succinic acid | C00042 | Citrate cycle (TCA cycle) | GC | C4H6O4 | 1.47 | 1.91 × 104 | 2.17 ↑ * |
| L-aspartate | C00049 | Cysteine and methionine metabolism | GC | C4H7NO4 | 1.29 | 4.77 × 103 | 2.02 ↑ * |
| L-glutamate | C00025 | Alanine, aspartate and glutamate metabolism | LC | C5H9NO4 | 8.24 | 1.44 × 105 | 1.52 ↑ |
| N-acetyl-glutamate | C01250 | Arginine biosynthesis | GC | C7H11NO4 | 1.84 | 2.72 × 104 | 3.37 ↑ * |
| Glutathione (GSH) | C00051 | Cysteine and methionine metabolism | ABC transporters | GC | C10H17N3O6S | 1.25 | 8.57 × 104 | 1.82 ↑ |
| L-gamma-glutamyl-L-valine | C03740 | Glutathione metabolism | LC | C10H18N2O5 | 1.34 | 3.31 × 106 | 1.34 ↑ |
| Malic acid | C00149 | Citrate cycle (TCA cycle) | GC | C4H6O5 | 1.13 | 2.28 × 104 | 2.62 ↓ |
| Fumaric acid | C00122 | Citrate cycle (TCA cycle) | GC | C4H4O4 | 1.49 | 3.51 × 103 | 1.52 ↓ |
| L-lactic acid | C00186 | Pyruvate metabolism | GC | C3H6O3 | 2.05 | 5.33 × 104 | 4.56 ↓ |
| L-Phenylalanine | C00079 | Phenylalanine metabolism | LC | C9H11NO2 | 5.94 | 2.58 × 104 | 3.24 ↓ |
| Phenyllactic acid | C05607 | Phenylalanine metabolism | LC | C9H10O3 | 1.15 | 8.02 × 108 | 1.61 ↓ |
| Aconitic acid | C00417 | Citrate cycle (TCA cycle) | GC | C6H6O6 | 1.19 | 2.58 × 104 | 1.54 ↑ |
| Citric acid | C00158 | Citrate cycle (TCA cycle) | GC | C6H8O7 | 2.62 | 3.81 × 106 | 5.75 ↓ * |
| L-methionine | C00073 | Cysteine and methionine metabolism | GC | C5H11NO2S | 1.17 | 1.14 × 103 | 1.69 ↓ |
| S-adenosyl-L-methionine (SAM) | C00019 | Cysteine and methionine metabolism | LC | C15H22N6O5S | 1.81 | 1.49 × 1010 | 16.66 ↓ * |
| S-ribosyl-L-homocysteine (SRH) | C03539 | Cysteine and methionine metabolism | LC | C9H17NO6S | 5.76 | 1.00 × 108 | 4.71 ↑ * |
| L-cystathionine | C02291 | Cysteine and methionine metabolism | GC | C7H14N2O4S | 2.40 | 9.01 × 108 | 6.85 ↓ * |
| L-homocysteine | C00155 | Cysteine and methionine metabolism | GC | C4H9NO2S | 2.03 | 1.44 × 105 | 1.73 ↓ |
| S-adenosyl-L-homocysteine (SAH) | C00021 | Cysteine and methionine metabolism | GC | C14H20N6O5S | 2.92 | 2.72 × 104 | 2.58 ↓ |
| Serine | C00065 | Cysteine and methionine metabolism | GC | C3H7NO3 | 1.58 | 6.93 × 105 | 0.42 ↓ |
| Homoserine | C00263 | Cysteine and methionine metabolism | GC | C4H9NO3 | 1.76 | 1.39 × 106 | 2.80 ↓ * |
| O-acetyl-L-serine | C00979 | Cysteine and methionine metabolism | GC | C5H9NO4 | 1.51 | 1.02 × 105 | 2.89 ↓ * |
| Creatine | C00300 | Glycine, serine and threonine metabolism | LC | C4H9N3O2 | 2.01 | 1.23 × 108 | 0.65 ↓ |
| D-fructose 2,6-bisphosphate | C00665 | Fructose and mannose metabolism | GC | C6H14O12P2 | 1.51 | 1.04 × 103 | 2.27 ↓ * |
| D-fructose | C02336 | Phosphotransferase system (PTS) Fructose and mannose metabolism | GC | C6H12O6 | 1.74 | 2.06 × 107 | 2.70 ↓ * |
| Cellobiose | C00185 | Phosphotransferase system (PTS) | GC | C12H22O11 | 1.73 | 1.40 × 106 | 1.59 ↓ |
| Galactose | C00984 | Galactose metabolism Phosphotransferase system (PTS) | GC | C6H12O6 | 1.48 | 5.98 × 106 | 2.08 ↓ * |
| Glucose-6-phosphate | C00092 | Phosphotransferase system (PTS) | GC | C6H13O9P | 1.35 | 1.41 × 104 | 1.93 ↓ |
| D-ribulose 5-phosphate | C00199 | Pentose phosphate pathway | GC | C5H11O8P | 1.09 | 3.31 × 103 | 1.61 ↓ |
| UDP-glucose | C00029 | Pyrimidine metabolism Pentose and glucuronate | LC | C15H24N2O17P2 | 1.53 | 3.97 × 105 | 3.80 ↓ * |
| D-glucose | C00031 | Glycolysis/Gluconeogenesis Phosphotransferase system (PTS) | LC | C6H12O6 | 2.04 | 2.79 × 105 | 2.33 ↓ * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, Y.; Li, Y.; Xu, T.; Zhao, H.; Zeng, M.; Liu, Z. Dissecting of the AI-2/LuxS Mediated Growth Characteristics and Bacteriostatic Ability of Lactiplantibacillus plantarum SS-128 by Integration of Transcriptomics and Metabolomics. Foods 2022, 11, 638. https://doi.org/10.3390/foods11050638
Qian Y, Li Y, Xu T, Zhao H, Zeng M, Liu Z. Dissecting of the AI-2/LuxS Mediated Growth Characteristics and Bacteriostatic Ability of Lactiplantibacillus plantarum SS-128 by Integration of Transcriptomics and Metabolomics. Foods. 2022; 11(5):638. https://doi.org/10.3390/foods11050638
Chicago/Turabian StyleQian, Yilin, Yuan Li, Tengteng Xu, Huijuan Zhao, Mingyong Zeng, and Zunying Liu. 2022. "Dissecting of the AI-2/LuxS Mediated Growth Characteristics and Bacteriostatic Ability of Lactiplantibacillus plantarum SS-128 by Integration of Transcriptomics and Metabolomics" Foods 11, no. 5: 638. https://doi.org/10.3390/foods11050638
APA StyleQian, Y., Li, Y., Xu, T., Zhao, H., Zeng, M., & Liu, Z. (2022). Dissecting of the AI-2/LuxS Mediated Growth Characteristics and Bacteriostatic Ability of Lactiplantibacillus plantarum SS-128 by Integration of Transcriptomics and Metabolomics. Foods, 11(5), 638. https://doi.org/10.3390/foods11050638






