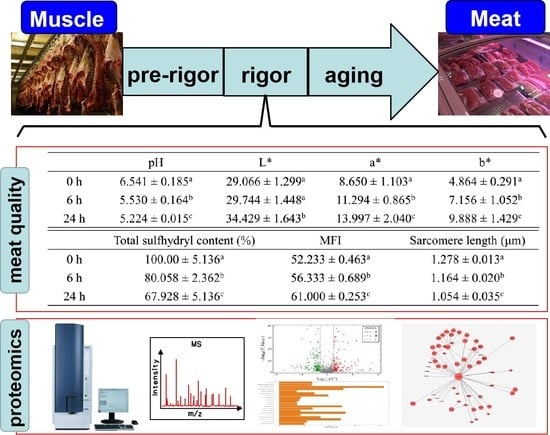
The Quality Changes and Proteomic Analysis of Cattle Muscle Postmortem during Rigor Mortis
Abstract
:1. Introduction
2. Materials and Methods
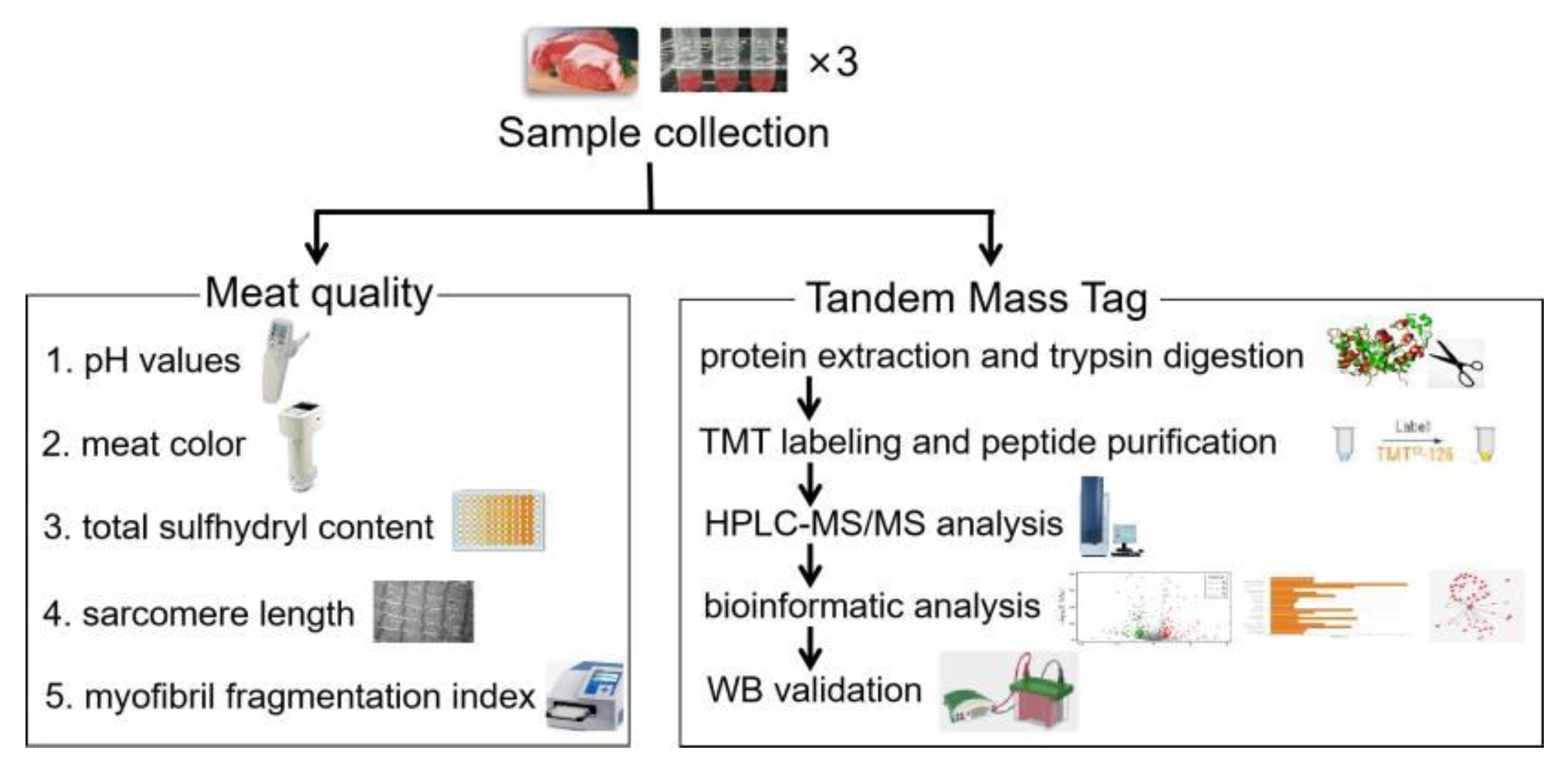
2.1. Animals and Muscle Sampling
2.2. pH Values
2.3. Meat Color
2.4. Total Sulfhydryl Content
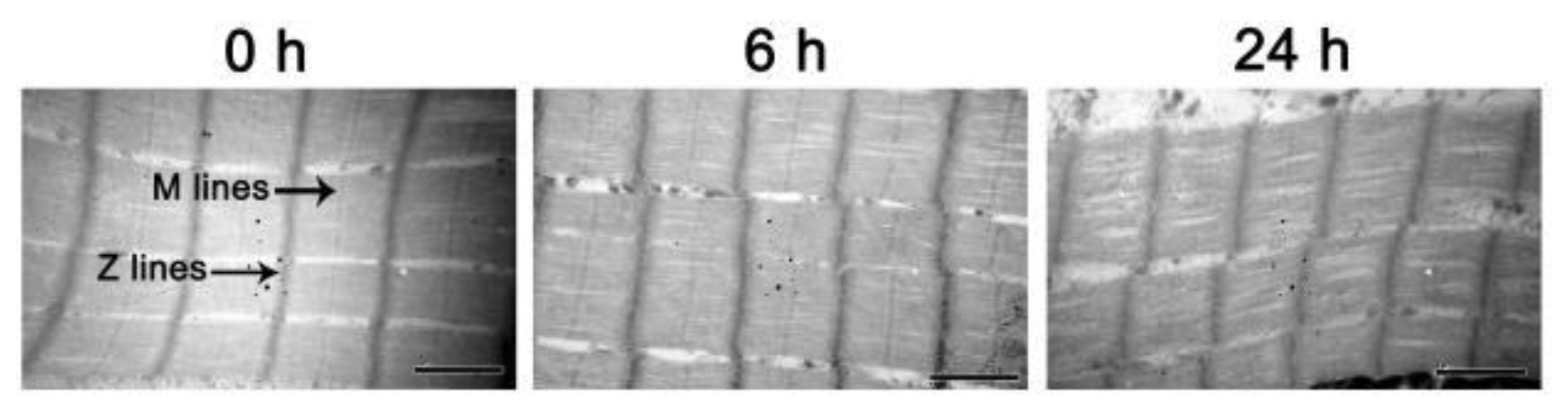
2.5. Sarcomere Length
2.6. Myofibril Fragmentation Index(MFI)
2.7. Trypsin Digestion and Tandem Mass Tag (TMT) Labeling
2.8. Peptide Purification and HPLC-MS/MS Analysis
2.9. Data Process and Bioinformatic Analysis
2.10. SDS-PAGE and Western Blotting
2.11. Statistical Analysis
3. Results and Discussion
3.1. pH and Meat Color
3.2. Total Sulfhydryl Content, Sarcomere Length and MFI
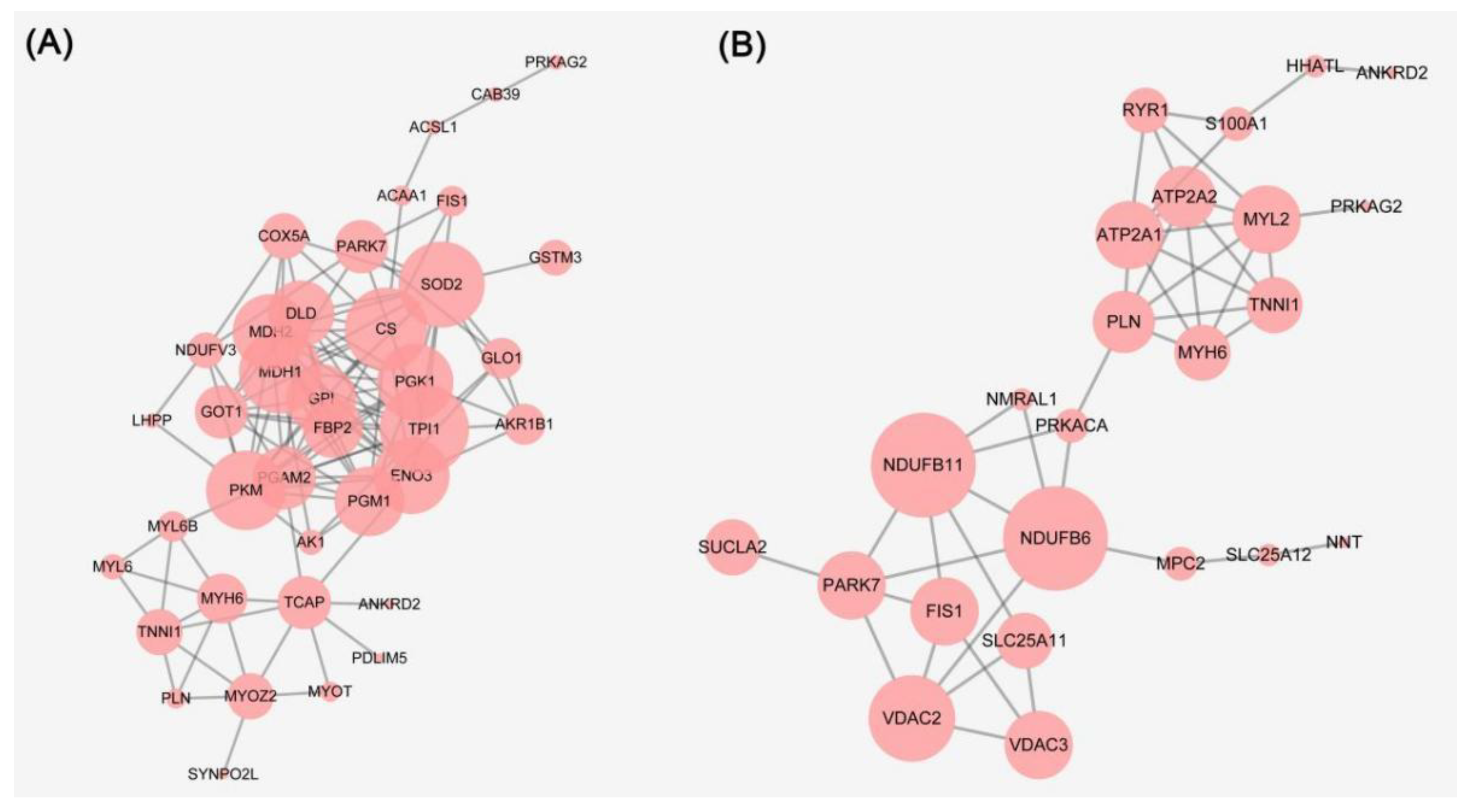
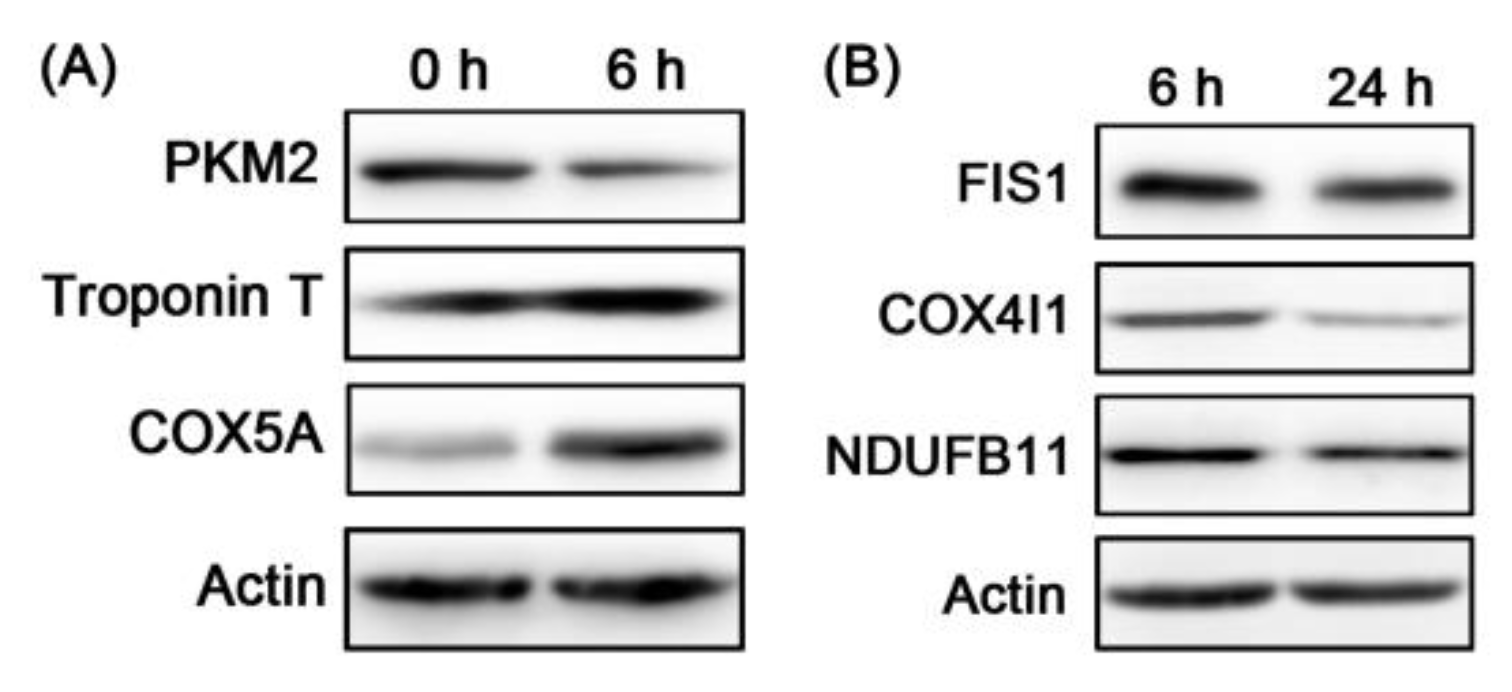
3.3. Bioinformatics Analysis of the Differential Proteins and WB Validation
3.4. Metabolic Enzymes
3.5. Mitochondrial Electron Transport Chain
3.6. Necroptosis and Ferroptosis
3.7. Structural Proteins
3.8. Potential Markers during Rigor Mortis
3.9. Meat Quality and Differentially Expressed Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hertzman, C.; Olsson, U.; Tornberg, E. The influence of high temperature, type of muscle and electrical stimulation on the course of rigor, ageing and tenderness of beef muscles. Meat Sci. 1993, 35, 119–141. [Google Scholar] [CrossRef]
- Ouali, A.; Herrera-Mendez, C.; Coulis, G.; Becila, S.; Boudjellal, A.; Aubry, L. Revisiting the conversion of muscle into meat and the underlying mechanisms. Meat Sci. 2006, 74, 44–58. [Google Scholar] [CrossRef]
- Li, C.; Shi, P.; Chang, X.; Xu, X.; Zhou, G. Tracing processes of rigor mortis and subsequent resolution of chicken breast muscle using a texture analyzer. J. Food Eng. 2010, 99, 388–391. [Google Scholar] [CrossRef]
- Matarneh, S.; England, E.; Scheffler, T.; Gerrard, D. The conversion of muscle to meat. In Lawrie´s Meat Science (eight edition); Woodhead Publishing Series in Food Science, Technology and Nutrition; Elsevier: Amsterdam, The Netherlands, 2017; pp. 159–185. [Google Scholar]
- Huang, C.; Hou, C.; Ijaz, M.; Yan, T.; Li, X.; Li, Y.; Zhang, D. Proteomics discovery of protein biomarkers linked to meat quality traits in post-mortem muscles: Current trends and future prospects: A review. Trends Food Sci. Technol. 2020, 105, 416–432. [Google Scholar] [CrossRef]
- Paredi, G.; Raboni, S.; Bendixen, E.; Almeida, A.; Mozzarelli, A. “Muscle to meat” molecular events and technological transformations: The proteomics insight. J. Proteom. 2012, 75, 4275–4289. [Google Scholar] [CrossRef]
- Poleti, M.D.; Moncau, C.T.; Silva-Vignato, B.; Rosa, A.F.; Lobo, A.R.; Cataldi, T.R.; Negrão, J.A.; Silva, S.L.; Eler, J.P.; Balieiro, J.C.D.C. Label-free quantitative proteomic analysis reveals muscle contraction and metabolism proteins linked to ultimate pH in bovine skeletal muscle. Meat Sci. 2018, 145, 209–219. [Google Scholar] [CrossRef]
- Bendixen, E. The use of proteomics in meat science. Meat Sci. 2005, 71, 138–149. [Google Scholar] [CrossRef]
- Gagaoua, M.; Bonnet, M.; Ellies-Oury, M.P.; De Koning, L.; Picard, B. Reverse phase protein arrays for the identification/validation of biomarkers of beef texture and their use for early classification of carcasses. Food Chem. 2018, 250, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Yu, Q.; Fu, Y.; Tian, X.; Jia, F.; Li, X.; Dai, R. Towards muscle-specific meat color stability of Chinese Luxi yellow cattle: A proteomic insight into post-mortem storage. J. Proteom. 2016, 147, 108–118. [Google Scholar] [CrossRef]
- Yu, Q.; Wu, W.; Tian, X.; Hou, M.; Dai, R.; Li, X. Unraveling proteome changes of Holstein beef M. semitendinosus and its relationship to meat discoloration during post-mortem storage analyzed by label-free mass spectrometry. J. Proteom. 2017, 154, 85–93. [Google Scholar] [CrossRef]
- Ding, Z.; Wei, Q.; Zhang, C.; Zhang, H.; Huang, F. Influence of oxidation on Heat Shock Protein 27 translocation, caspase-3 and calpain activities and myofibrils degradation in postmortem beef muscles. Food Chem. 2020, 340, 127914. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, D.; Xu, X.; Xu, W. Comparative proteomic analysis of proteins associated with water holding capacity in goose muscles. Food Res. Int. 2018, 116, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Malheiros, J.; Braga, C.; Grove, R.; Ribeiro, F.; Calkins, C.; Adamec, J.; Chardulo, L. Influence of oxidative damage to proteins on meat tenderness using a proteomics approach. Meat Sci. 2019, 148, 64–71. [Google Scholar] [CrossRef]
- Gagaoua, M.; Bonnet, M.; De Koning, L.; Picard, B. Reverse phase protein array for the quantification and validation of protein biomarkers of beef qualities: The case of meat color from Charolais breed. Meat Sci. 2018, 145, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Sentandreu, M.A.; Coulis, G.; Ouali, A. Role of muscle endopeptidases and their inhibitors in meat tenderness. Trends Food Sci. Technol. 2002, 13, 400–421. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, D.; Huang, W.; Liu, F.; Zhu, Y.; Xu, W.; Cao, J. Apoptosis during postmortem conditioning and its relationship to duck meat quality. Food Chem. 2013, 138, 96–100. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Xia, C.; He, Y.; Pan, D.; Cao, J.; Sun, Y.; Zeng, X. Proteomic responses to oxidative damage in meat from ducks exposed to heat stress. Food Chem. 2019, 295, 129–137. [Google Scholar] [CrossRef]
- Alvarado, C.Z.; Sams, A.R. Rigor mortis development in Turkey breast muscle and the effect of electrical stunning. Poult. Sci. 2000, 79, 1694–1698. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yu, Q.; Han, L.; Zhang, Y.; Tian, X.; Zhao, S. Effects of proteome changes on the tenderness of yak rumen smooth muscle during postmortem storage based on the label-free mass spectrometry. Food Res. Int. 2019, 116, 1336–1343. [Google Scholar] [CrossRef]
- Beldarrain, L.R.; Aldai, N.; Picard, B.; Sentandreu, E.; Navarro, J.L.; Sentandreu, M.A. Use of liquid isoelectric focusing (OFFGEL) on the discovery of meat tenderness biomarkers. J. Proteom. 2018, 183, 25–33. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, B.; Huang, Y.; Chen, L.; Bao, P.; Fang, H.; Zhou, C. L-arginine and L-lysine degrade troponin-T, and L-arginine dissociates actomyosin: Their roles in improving the tenderness of chicken breast. Food Chem. 2020, 318, 126516. [Google Scholar] [CrossRef]
- Merino, F.; Pospich, S.; Funk, J.; Wagner, T.; Raunser, S. Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat. Struct. Mol. Biol. 2018, 25, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, X.; Li, Z.; Li, M.; Zhu, J.; Zhang, D. Changes in degradation and phosphorylation level of titin in three ovine muscles during postmortem. Int. J. Food Sci. Technol. 2018, 53, 913–920. [Google Scholar] [CrossRef]
- Yu, L.H.; Lee, E.S.; Jeong, J.Y.; Choi, J.H.; Kim, C.J. Effects of Post-mortem Temperature on the Physicochemical Properties of Hot-boned Chicken Breast Muscles. Food Sci. Anim. Resour. 2009, 29, 55–61. [Google Scholar] [CrossRef]
- Ertbjerg, P.; Puolanne, E. Muscle structure, sarcomere length and influences on meat quality: A review. Meat Sci. 2017, 132, 139–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornforth, D.; Pearson, A.; Merkel, R. Relationship of mitochondria and sarcoplasmic reticulum to cold shortening. Meat Sci. 1980, 4, 103–121. [Google Scholar] [CrossRef]
- Alquraishi, M.; Puckett, D.L.; Alani, D.S.; Humidat, A.S.; Bettaieb, A. Pyruvate kinase M2: A simple molecule with complex functions. Free Radic. Biol. Med. 2019, 143, 176–192. [Google Scholar] [CrossRef]
- Marsh, B.B. Rigor mortis in beef. J. Sci. Food Agric. 2010, 5, 70–75. [Google Scholar] [CrossRef]
- Gagaoua, M.; Couvreur, S.; Bec, G.L.; Aminot, G.; Picard, B. Associations among protein biomarkers and pH and color traits in longissimus thoracis and rectus abdominis muscles in protected designation of origin maine-Anjou cull cows. J. Agric. Food Chem. 2017, 65, 3569–3580. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Feng, D.; Xiao, J.; Wu, F.; Zuo, J.J. Studies on meat color, myoglobin content, enzyme activities, and genes associated with oxidative potential of pigs slaughtered at different growth stages. Asian-Australas. J. Anim. Sci. 2017, 30, 1739–1750. [Google Scholar] [CrossRef] [Green Version]
- Belskie, K.M.; Buiten, C.; Ramanathan, R.; Mancini, R.A. Reverse electron transport effects on NADH formation and metmyoglobin reduction. Meat Sci. 2015, 105, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Degterev, A.; Huang, Z.; Boyce, M.; Li, Y.; Jagtap, P.; Mizushima, N.; Cuny, G.D.; Mitchison, T.J.; Moskowitz, M.A.; Yuan, J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005, 1, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Ropka-Molik, K.; Żukowski, K.; Eckert, R.; Gurgul, A.; Piórkowska, K.; Oczkowicz, M. Comprehensive analysis of the whole transcriptomes from two different pig breeds using RNA-Seq method. Anim. Genet. 2015, 45, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhang, Y.; Wu, J.; Qiao, M.; Mei, S. Proteomic and lipidomic analyses reveal saturated fatty acids, phosphatidylinositol, phosphatidylserine, and associated proteins contributing to intramuscular fat deposition. J. Proteom. 2021, 241, 104235. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Luo, M.; Zhang, K.; Zhang, J.; Chen, W. Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun. 2020, 11, 433. [Google Scholar] [CrossRef] [Green Version]
- Siragusa, M.; Sessa, W.C. Telmisartan exerts pleiotropic effects in endothelial cells and promotes endothelial cell quiescence and survival. Arter. Thromb. Vasc. Biol. 2013, 33, 1852–1860. [Google Scholar] [CrossRef] [Green Version]
- Zuo, H.; Han, L.; Yu, Q.; Niu, K.; Zhao, S.; Shi, H. Proteome changes on water-holding capacity of yak longissimus lumborum during postmortem aging. Meat Sci. 2016, 121, 409–419. [Google Scholar] [CrossRef]
- Gustafson, P.; Lin, K.C.; Xu, J.; Kamara, D.; Geng, T.; Gyenai, K.; Reed, K.M.; Smith, E.J. DNA sequence and haplotype variation in two candidate genes for dilated cardiomyopathy in the turkey Meleagris gallopavo. Genome 2015, 50, 463–469. [Google Scholar]
- Krishnamoorthy, S.; Jin, R.; Cai, Y.; Maddipati, K.R.; Nie, D.; Pagès, G.; Tucker, S.C.; Honn, K.V. 12-Lipoxygenase and the regulation of hypoxia-inducible factor in prostate cancer cells. Exp. Cell Res. 2010, 316, 1706–1715. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Lei, M.; Xiong, Y. Molecular Characterization and Different Expression Patterns of the Muscle Ankyrin Repeat Protein (MARP) Family During Porcine Skeletal Muscle Development in vitro and in vivo. Anim. Biotechnol. 2011, 22, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Pallavicini, A.; Kojić, S.; Bean, C.; Vainzof, M.; Salamon, M.; Ievolella, C.; Bortoletto, G.; Pacchioni, B.; Zatz, M.; Lanfranchi, G. Characterization of Human Skeletal Muscle Ankrd2. Biochem. Biophys. Res. Commun. 2001, 285, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Dong, X.; Fan, B.; Liu, B. The Association of ANKRD2 with Loin Depth and Muscle Firmness in Pigs. J. Anim. Vet. Adv. 2011, 10, 1462–1468. [Google Scholar] [CrossRef] [Green Version]
- Rosenvold, K.; Petersen, J.; Laerke, H.; Jensen, S.; Therkildsen, M.; Karlsson, A.; Moller, H.; Andersen, H. Muscle glycogen stores and meat quality as affected by strategic finishing feeding of slaughter pigs. J. Anim. Sci. 2001, 79, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.; Joseph, P. Myoglobin chemistry and meat color. Annu. Rev. Food Sci. Technol. 2013, 4, 79–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Li, Z.; Everaert, N.; Lametsch, R.; Zhang, D. Quantitative phosphoproteomic analysis of ovine muscle with different postmortem glycolytic rates. Food Chem. 2019, 280, 203–209. [Google Scholar] [PubMed]
- Wu, W.; Gao, X.; Dai, Y.; Fu, Y.; Li, X.; Dai, R. Post-mortem changes in sarcoplasmic proteome and its relationship to meat color traits in M. semitendinosus of Chinese Luxi yellow cattle. Food Res. Int. 2015, 72, 98–105. [Google Scholar] [CrossRef]
| pH | L* | a* | b* | |
|---|---|---|---|---|
| 0 h | 6.541 ± 0.185 a | 29.066 ± 1.299 a | 8.650 ± 1.103 a | 4.864 ± 0.291 a |
| 6 h | 5.530 ± 0.164 b | 29.744 ± 1.448 a | 11.294 ± 0.865 b | 7.156 ± 1.052 b |
| 24 h | 5.224 ± 0.015 c | 34.429 ± 1.643 b | 13.997 ± 2.040 c | 9.888 ± 1.429 c |
| Total Sulfhydryl Content (%) | MFI | Sarcomere Length (μm) | |
|---|---|---|---|
| 0 h | 100.00 ± 5.136 a | 52.233 ± 0.463 a | 1.278 ± 0.013 a |
| 6 h | 80.058 ± 2.362 b | 56.333 ± 0.689 b | 1.164 ± 0.020 b |
| 24 h | 67.928 ± 5.136 c | 61.000 ± 0.253 c | 1.054 ± 0.035 c |
| Term | Description | p Value | |
|---|---|---|---|
| Biological Process | GO:0046031 | ADP metabolic process | 6.10 × 10−9 |
| GO:0006757 | ATP generation from ADP | 3.02 × 10−8 | |
| GO:0009132 | Nucleoside diphosphate metabolic process | 1.02 × 10−8 | |
| GO:0006165 | Nucleoside diphosphate phosphorylation | 2.03 × 10−9 | |
| GO:0046939 | Nucleotide phosphorylation | 2.03 × 10−9 | |
| GO:0046434 | Organophosphate catabolic process | 3.67 × 10−8 | |
| GO:0009135 | Purine nucleoside diphosphate metabolic process | 6.10 × 10−9 | |
| GO:0009179 | Purine ribonucleoside diphosphate metabolic process | 6.10 × 10−9 | |
| GO:0042866 | Pyruvate biosynthetic process | 3.02 × 10−8 | |
| GO:0009185 | Ribonucleoside diphosphate metabolic process | 6.10 × 10−9 | |
| Cellular Component | GO:0005903 | Brush border | 0.043 |
| GO:1990584 | Cardiac troponin complex | 0.037 | |
| GO:0043292 | Contractile fiber | 0.009 | |
| GO:0044449 | Contractile fiber part | 0.008 | |
| GO:0098850 | Extrinsic component of synaptic vesicle membrane | 0.020 | |
| GO:0097452 | GAIT complex | 0.037 | |
| GO:0031674 | I band | 0.037 | |
| GO:0030016 | Myofibril | 0.009 | |
| GO:0030017 | Sarcomere | 0.006 | |
| GO:0005861 | Troponin complex | 0.015 | |
| Molecular Function | GO:0051373 | FATZ binding | 4.58 × 10−5 |
| GO:0016866 | Intramolecular transferase activity | 2.14 × 10−4 | |
| GO:0016868 | Intramolecular transferase activity, phosphotransferases | 0.002 | |
| GO:0000287 | Magnesium ion binding | 0.006 | |
| GO:0004784 | Superoxide dismutase activity | 0.007 | |
| GO:0016532 | Superoxide dismutase copper chaperone activity | 0.007 | |
| GO:0031433 | Telethonin binding | 0.002 | |
| GO:0016740 | Transferase activity | 9.75 × 10−4 | |
| GO:0016769 | Transferase activity, transferring nitrogenous groups | 0.001 | |
| GO:0009041 | Uridylate kinase activity | 0.007 |
| Term | Description | p Value | |
|---|---|---|---|
| Biological Process | GO:0098656 | Anion transmembrane transport | 2.42 × 10−5 |
| GO:0006820 | Anion transport | 9.35 × 10−5 | |
| GO:1905039 | Carboxylic acid transmembrane transport | 3.52 × 10−4 | |
| GO:0015893 | Drug transport | 9.31 × 10−6 | |
| GO:0051234 | Establishment of localization | 3.68 × 10−4 | |
| GO:0034220 | Ion transmembrane transport | 6.68 × 10−4 | |
| GO:0006811 | Ion transport | 2.37 × 10−5 | |
| GO:0021675 | Nerve development | 3.52 × 10−4 | |
| GO:1903825 | Organic acid transmembrane transport | 3.52 × 10−4 | |
| GO:0006810 | Transport | 3.42 × 10−4 | |
| Cellular Component | GO:0031975 | Envelope | 4.82 × 10−4 |
| GO:0016021 | Integral component of membrane | 3.94 × 10−10 | |
| GO:0031301 | Integral component of organelle membrane | 5.38 × 10−4 | |
| GO:0031224 | Intrinsic component of membrane | 5.74 × 10−10 | |
| GO:0098573 | Intrinsic component of mitochondrial membrane | 5.38 × 10−4 | |
| GO:0044425 | Membrane part | 1.88 × 10−4 | |
| GO:0005740 | Mitochondrial envelope | 3.47 × 10−4 | |
| GO:0031966 | Mitochondrial membrane | 3.52 × 10−4 | |
| GO:0031967 | Organelle envelope | 4.82 × 10−4 | |
| GO:0031090 | Organelle membrane | 1.49 × 10−4 | |
| Molecular Function | GO:0022804 | Active transmembrane transporter activity | 6.62 × 10−5 |
| GO:0008509 | Anion transmembrane transporter activity | 7.26 × 10−7 | |
| GO:0046943 | Carboxylic acid transmembrane transporter activity | 3.52 × 10−4 | |
| GO:0015238 | Drug transmembrane transporter activity | 3.52 × 10−4 | |
| GO:0015318 | Inorganic molecular entity transmembrane transporter activity | 8.75 × 10−6 | |
| GO:0015075 | Ion transmembrane transporter activity | 8.75 × 10−6 | |
| GO:0008514 | Organic anion transmembrane transporter activity | 2.42 × 10−5 | |
| GO:0015291 | Secondary active transmembrane transporter activity | 2.42 × 10−5 | |
| GO:0022857 | Transmembrane transporter activity | 3.15 × 10−6 | |
| GO:0005215 | Transporter activity | 6.38 × 10−6 |
| Pathway | Pathway Name | p Value |
|---|---|---|
| map00220 | Arginine biosynthesis | 0.037 |
| map01230 | Biosynthesis of amino acids | 2.39 × 10−5 |
| map01200 | Carbon metabolism | 7.47 × 10−6 |
| map05204 | Chemical carcinogenesis | 0.005 |
| map00270 | Cysteine and methionine metabolism | 0.006 |
| map00982 | Drug metabolism-cytochrome P450 | 0.009 |
| map00051 | Fructose and mannose metabolism | 0.009 |
| map00052 | Galactose metabolism | 0.037 |
| map00480 | Glutathione metabolism | 0.006 |
| map00010 | Glycolysis/Gluconeogenesis | 7.77 × 10−9 |
| map04910 | Insulin signaling pathway | 0.029 |
| map04213 | Longevity regulating pathway-multiple species | 0.009 |
| map01100 | Metabolic pathways | 3.48 × 10−7 |
| map00980 | Metabolism of xenobiotics by cytochrome P450 | 0.015 |
| map00030 | Pentose phosphate pathway | 0.004 |
| map04146 | Peroxisome | 0.023 |
| map00360 | Phenylalanine metabolism | 0.037 |
| map00620 | Pyruvate metabolism | 0.001 |
| map00500 | Starch and sucrose metabolism | 0.032 |
| map00750 | Vitamin B6 metabolism | 0.007 |
| Pathway | Pathway Name | p Value |
|---|---|---|
| map05010 | Alzheimer’s disease | 0.013 |
| map04020 | Calcium signaling pathway | 8.00 × 10−5 |
| map04022 | cGMP-PKG signaling pathway | 4.70 × 10−4 |
| map05166 | HTLV-I infection | 0.002 |
| map05016 | Huntington’s disease | 0.002 |
| map04657 | IL-17 signaling pathway | 0.044 |
| map04211 | Longevity regulating pathway | 0.044 |
| map05144 | Malaria | 0.027 |
| map04217 | Necroptosis | 0.039 |
| map00190 | Oxidative phosphorylation | 0.006 |
| map05012 | Parkinson’s disease | 6.80 × 10−5 |
| map04742 | Taste transduction | 0.005 |
| map00130 | Ubiquinone and other terpenoid–quinone biosynthesis | 0.014 |
| Accession | Gene Names | TMT (6/0 h) | WB (6/0 h) | TMT (24/6 h) | WB (24/6 h) | ||||
|---|---|---|---|---|---|---|---|---|---|
| FC | p Value | FC | p Value | FC | p Value | FC | p Value | ||
| A5D984 | PKM | 0.64 | 0 | 0.64 | 0.0056 | ||||
| Q8MKH6-2 | TNNT1 | 1.47 | 2.15 × 10−33 | 1.57 | 0.0024 | ||||
| P00426 | COX5A | 2.28 | 0.0001 | 1.72 | 0.0017 | ||||
| Q3T0I5 | FIS1 | 0.78 | 2.58 × 10−6 | 0.65 | 0.0031 | ||||
| P00423 | COX4I1 | 0.49 | 3.34 × 10−36 | 0.61 | 0.0318 | ||||
| Q8HXG5 | NDUFB11 | 0.87 | 0.0387 | 0.66 | 0.0216 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Z.; Wei, Q.; Liu, C.; Zhang, H.; Huang, F. The Quality Changes and Proteomic Analysis of Cattle Muscle Postmortem during Rigor Mortis. Foods 2022, 11, 217. https://doi.org/10.3390/foods11020217
Ding Z, Wei Q, Liu C, Zhang H, Huang F. The Quality Changes and Proteomic Analysis of Cattle Muscle Postmortem during Rigor Mortis. Foods. 2022; 11(2):217. https://doi.org/10.3390/foods11020217
Chicago/Turabian StyleDing, Zhenjiang, Qichao Wei, Chunmei Liu, Hong Zhang, and Feng Huang. 2022. "The Quality Changes and Proteomic Analysis of Cattle Muscle Postmortem during Rigor Mortis" Foods 11, no. 2: 217. https://doi.org/10.3390/foods11020217
APA StyleDing, Z., Wei, Q., Liu, C., Zhang, H., & Huang, F. (2022). The Quality Changes and Proteomic Analysis of Cattle Muscle Postmortem during Rigor Mortis. Foods, 11(2), 217. https://doi.org/10.3390/foods11020217