Reduction of Pathogens in Feces and Lymph Nodes Collected from Beef Cattle Fed Lactobacillus salivarius (L28), Lactobacillus acidophilus (NP51) and Propionibacterium freudenreichii (NP28), Commercially Available Direct-Fed Microbials
Abstract
:1. Introduction
2. Materials and Methods
2.1. Treatment Groups—Cattle Feed Yards
2.2. Sample Collection
2.3. Fecal Sample Analysis
2.4. Lymph Node Sample Processing and Salmonella Detection
2.5. Statistical Analysis
3. Results
3.1. Fecal Samples
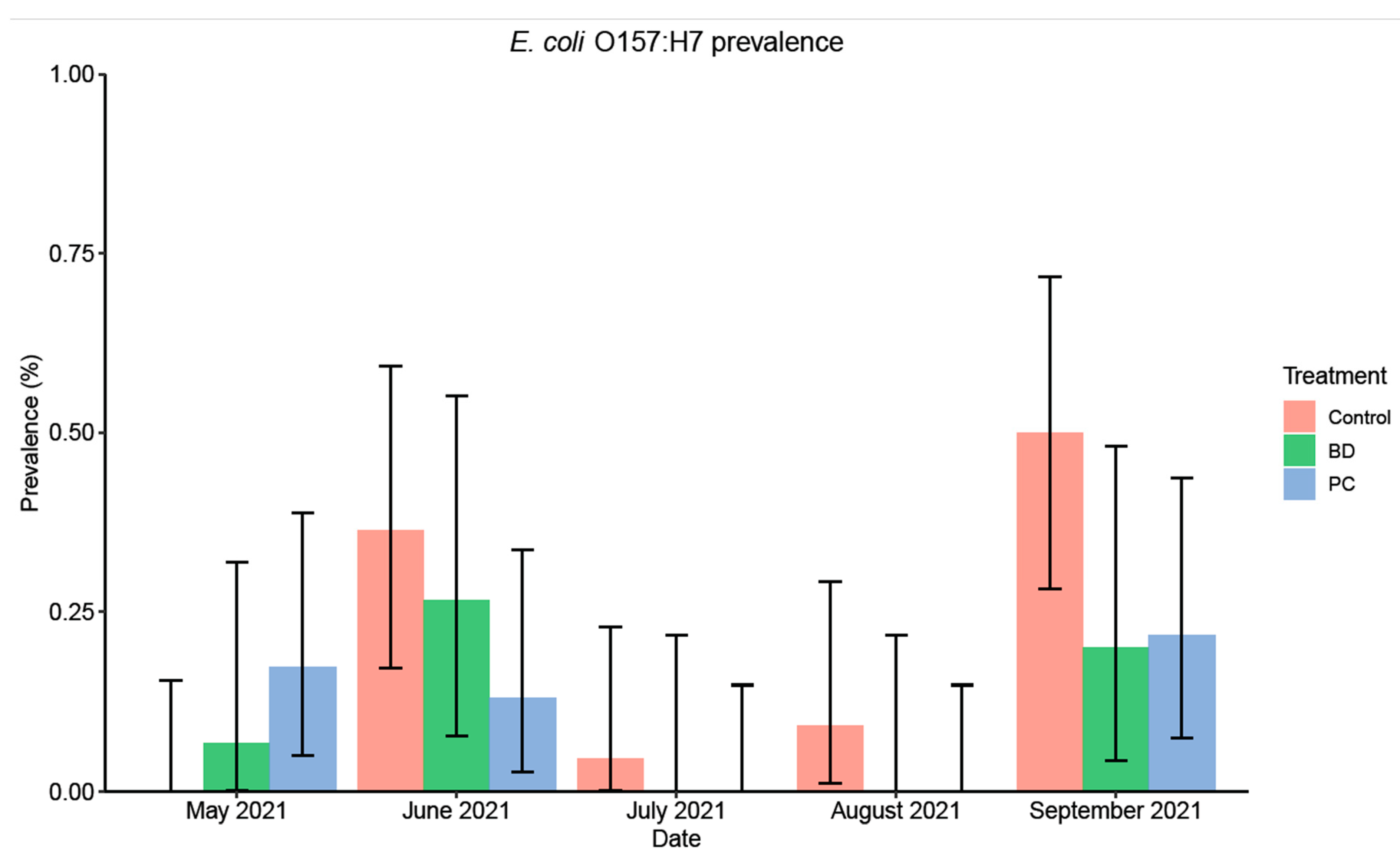
3.1.1. E. coli O157:H7 Prevalence and Concentration
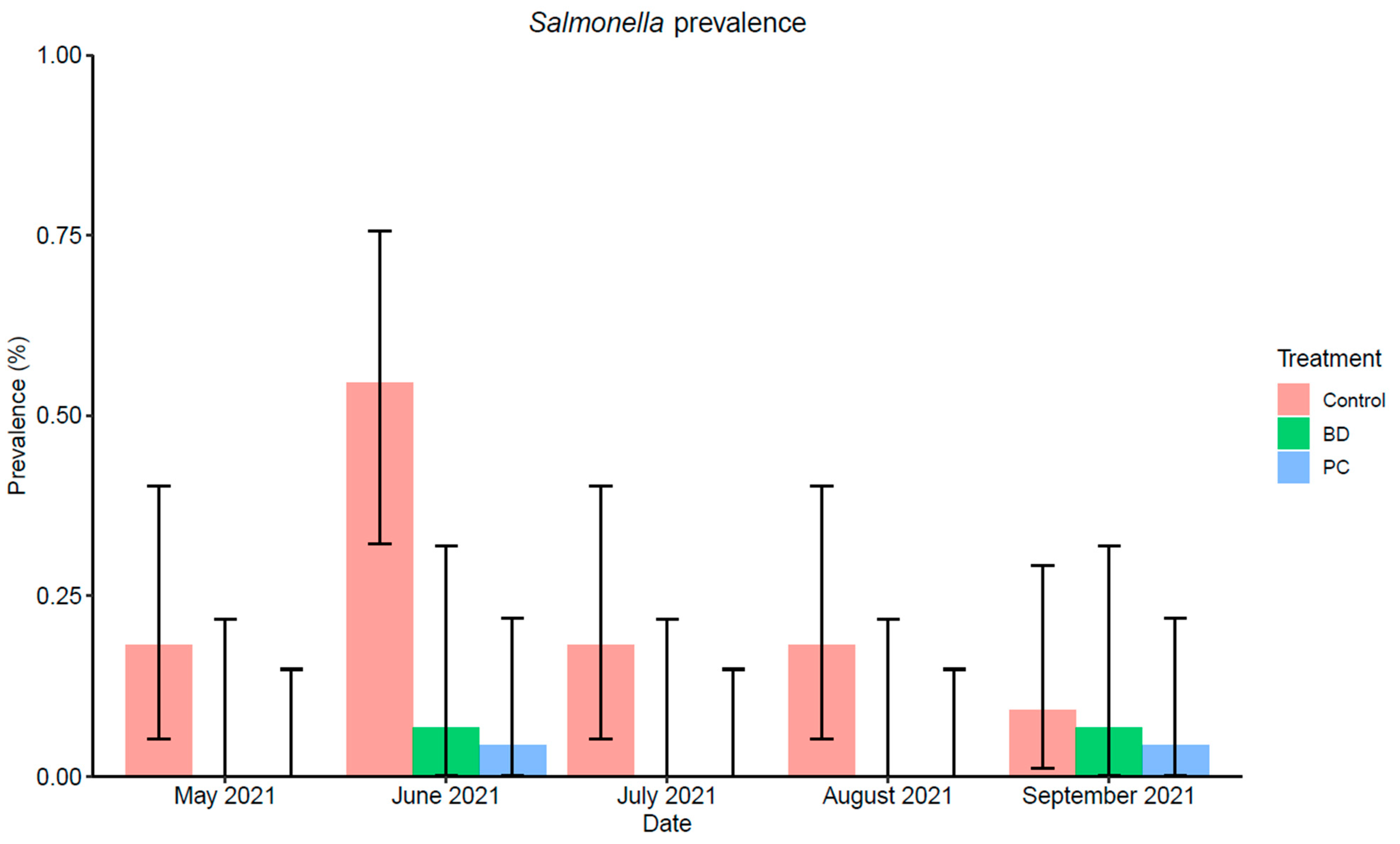
3.1.2. Salmonella Prevalence
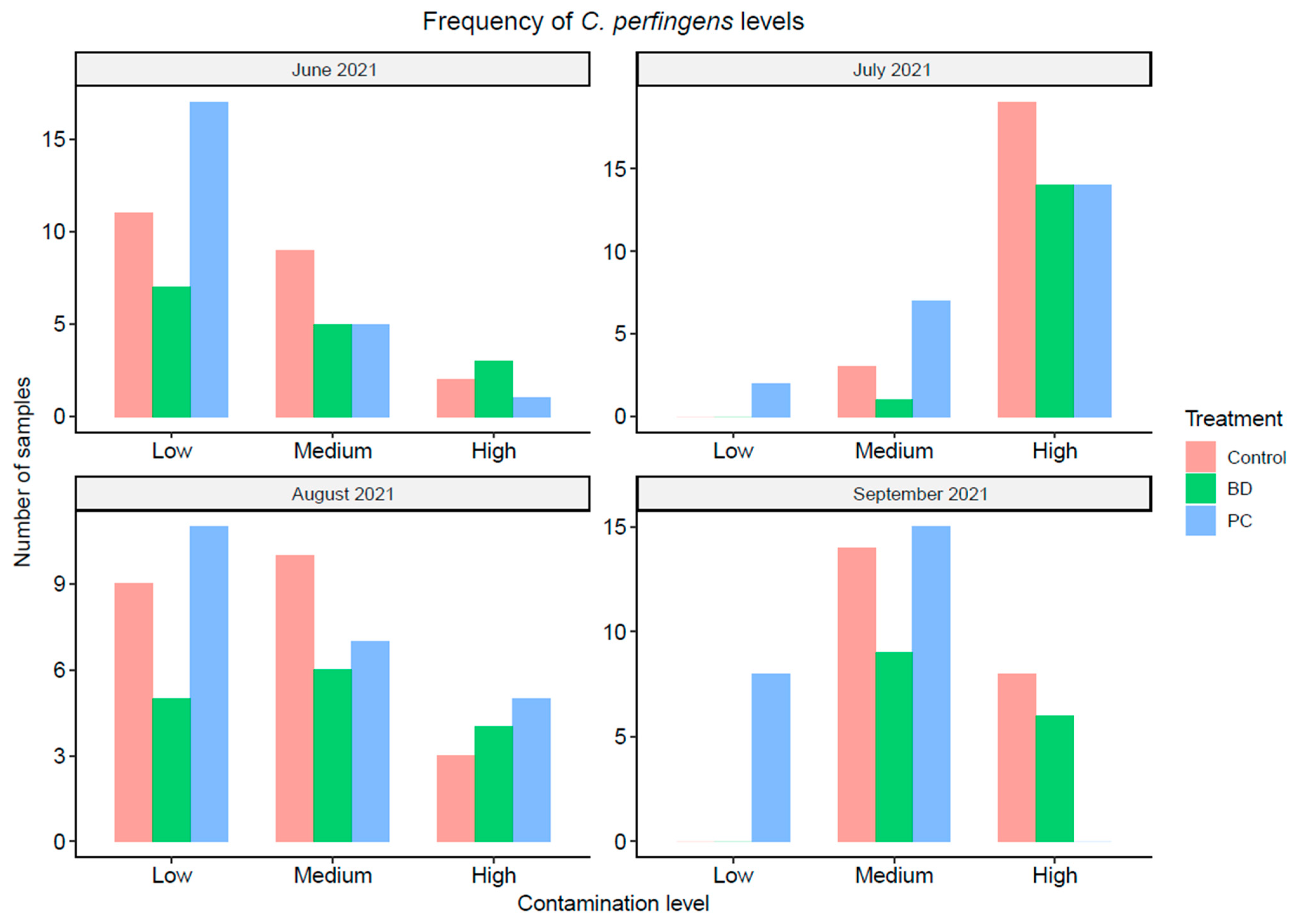
3.1.3. Clostridium Perfringens Concentration
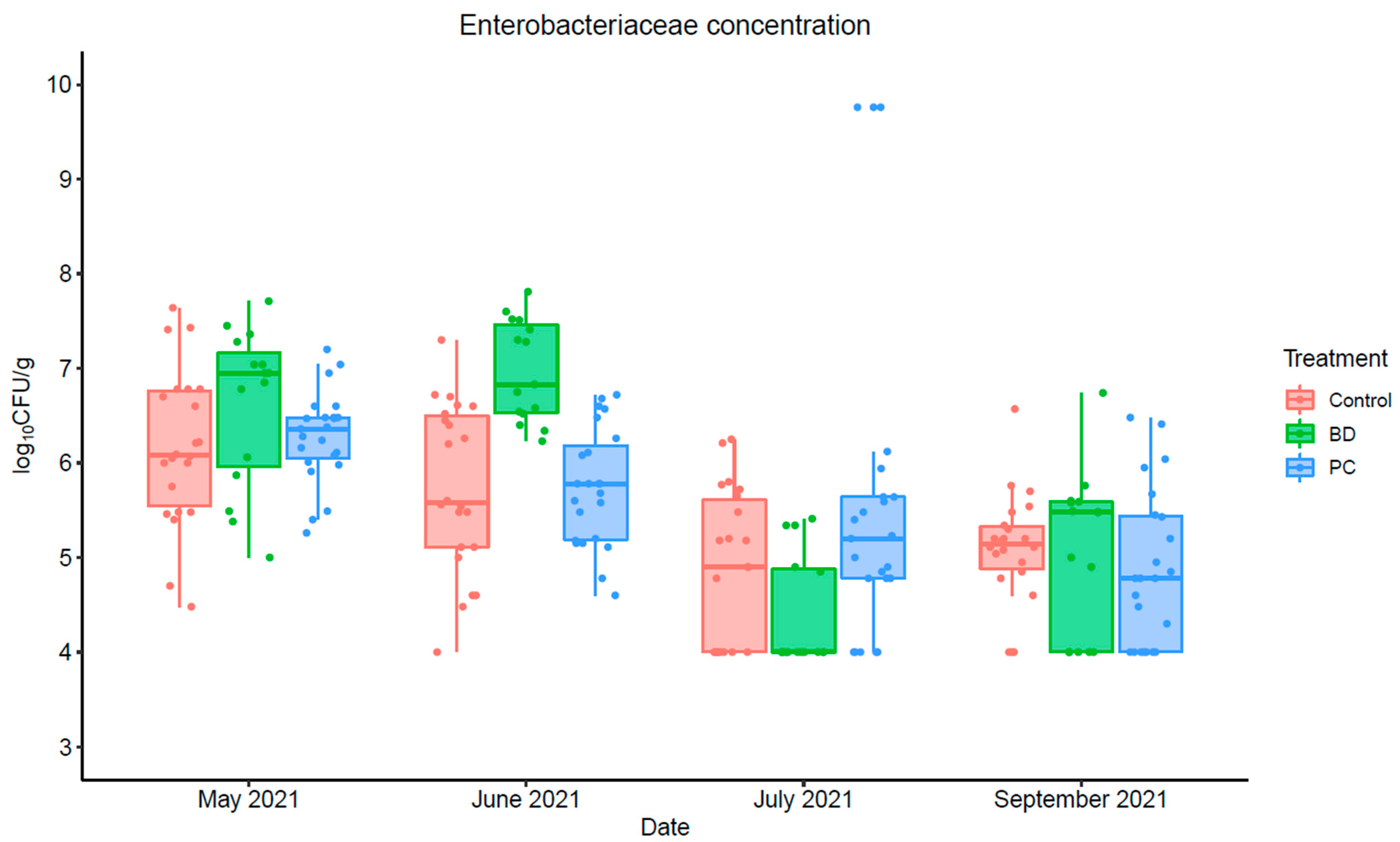
3.1.4. Enterobacteriaceae Concentration
3.2. Peripheral Lymph Node Samples
Salmonella Prevalence
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Interagency Food Safety Analytics Collaboration [IFSAC]. Foodborne Illness Source Attribution Estimates for 2019 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter Using Multi-Year Outbreak Surveillance Data, United States; GA and D.C: U.S. Department of Health and Human Services’ Centers for Disease Control and Prevention and U.S. Food and Drug Administration, U.S. Department of Agriculture’s Food Safety and Inspection Service 2021. Available online: https://www.cdc.gov/foodsafety/ifsac/pdf/P19-2019-report-TriAgency-508.pdf (accessed on 2 September 2022).
- Scallan, E.; Hoekstra, R.M.; Mahon, B.E.; Jones, T.F.; Griffin, P.M. An assessment of the human health impact of seven leading foodborne pathogens in the United States using disability adjusted life years. Epidemiol. Infect. 2015, 143, 2795–2804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Office of Disease Prevention and Health Promotion. Healthy People 2030 Framework. Available online: https://health.gov/healthypeople/about/healthy-people-2030-framework (accessed on 15 July 2022).
- Donkersgoed, J.V.; Bohaychuk, V.; Besser, T.; Song, X.M.; Wagner, B.; Hancock, D.; Renter, D.; Dargatz, D. Occurrence of foodborne bacteria in Alberta feedlots. Can. Vet. J. 2009, 50, 166–172. [Google Scholar] [PubMed]
- Brashears, M.M.; Chaves, B.D. The diversity of beef safety: A global reason to strengthen our current systems. Meat Sci. 2017, 132, 59–71. [Google Scholar] [CrossRef]
- Stephens, T.P.; Loneragan, G.H.; Thompson, T.W.; Sridhara, A.; Branham, L.A.; Pitchiah, S.; Brashears, M.M. Distribution of Escherichia coli O157 and Salmonella on hide surfaces, the oral cavity, and in feces of feedlot cattle. J. Food Prot. 2007, 70, 1346–1349. [Google Scholar] [CrossRef] [PubMed]
- Arthur, T.M.; Keen, J.E.; Bosilevac, J.M.; Brichta-Harhay, D.M.; Kalchayanand, N.; Shackelford, S.D.; Wheeler, T.L.; Nou, X.; Koohmaraie, M. Longitudinal study of Escherichia coli O157:H7 in a beef cattle feedlot and role of high-level shedders in hide contamination. Appl. Environ. Microbiol. 2009, 75, 6515–6523. [Google Scholar] [CrossRef] [Green Version]
- Berry, E.D.; Wells, J.E. Chapter 4—Escherichia coli O157:H7: Recent advances in research on occurrence, transmission, and control in cattle and the production environment. Adv. Food Nutr. Res. 2010, 60, 67–117. [Google Scholar] [CrossRef] [PubMed]
- Callaway, T.R.; Edrington, T.S.; Brabban, A.D.; Keen, J.E.; Anderson, R.C.; Rossman, M.L.; Engler, M.J.; Genovese, K.J.; Gwartney, B.L.; Reagan, J.O.; et al. Fecal prevalence of Escherichia coli O157, Salmonella, Listeria, and bacteriophage infecting E. coli O157:H7 in feedlot cattle in the southern plains region of the United States. Foodborne Pathog. Dis. 2006, 3, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Tschirdewahn, B.; Notermans, S.; Wernars, K.; Untermann, F. The presence of enterotoxigenic Clostridium perfringens strains in faeces of various animals. Int. J. Food Microbiol. 1991, 14, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Hamza, D.; Dorgham, S.M.; Elhariri, M.; Elhelw, R.; Ismael, E. New insight of apparently healthy animals as a potential reservoir for Clostridium perfringens: A public health implication. J. Vet. Res. 2018, 62, 457–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uzal, F.A.; McClane, B.A.; Cheung, J.K.; Theoret, J.; Garcia, J.P.; Moore, R.J.; Rood, J.I. Animal models to study the pathogenesis of human and animal Clostridium perfringens infections. Vet. Microbiol. 2015, 179, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Interagency Food Safety Analytics Collaboration [IFSAC]. Foodborne Illness Source Attribution Estimates for 2013 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter Using Multi-Year Outbreak Surveillance Data, United States; GA and D.C.: U.S. Department of Health and Human Services, CDC, FDA, USDA-FSIS, 2017. Available online: https://www.cdc.gov/foodsafety/pdfs/IFSAC-2013FoodborneillnessSourceEstimates-508.pdf (accessed on 2 September 2022).
- Wheeler, T.L.; Kalchayan, N.; Bosilevac, J.M. Pre- and post-harvest interventions to reduce pathogen contamination in the U.S. beef industry. Meat Sci. 2014, 98, 372–382. [Google Scholar] [CrossRef] [Green Version]
- Loneragan, G.H.; Brashears, M.M. Pre-harvest interventions to reduce carriage of E. coli O157 by harvest-ready feedlot cattle. Meat Sci. 2005, 71, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Sargeant, J.M.; Amezcua, M.R.; Rajic, A.; Waddell, L. Pre-harvest interventions to reduce the shedding of E. coli O157 in the faeces of weaned domestic ruminants: A systematic review. Zooneses Public Health 2007, 54, 260–277. [Google Scholar] [CrossRef]
- Stephens, T.P.; Loneragan, G.H.; Chichester, L.M.; Brashears, M.M. Prevalence and enumeration of Escherichia coli O157 in steers receiving various strains of Lactobacillus-based direct-fed microbials. J. Food Prot. 2007, 70, 1252–1255. [Google Scholar] [CrossRef]
- Cull, C.A.; Paddocka, Z.D.; Nagarajaa, T.G.; Bello, N.M.; Babcockc, A.H.; Renter, D.G. Efficacy of a vaccine and a direct-fed microbial against fecal shedding of Escherichia coli O157:H7 in a randomized pen-level field trial of commercial feedlot cattle. Vaccine 2012, 30, 6210–6215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brashears, M.M.; Amezquita, A.; Jaroni, D. Lactic acid bacteria and their uses in animal feeding to improve food safety. Adv. Food Nutr. Res. 2005, 50, 2–25. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Compliance Policy Guideline Sec. 689.100 Direct-Fed Microbial Products. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/cpg-sec-689100-direct-fed-microbial-products (accessed on 15 July 2022).
- Opheim, T.L. The Effects of a Novel Direct-Fed Microbial on Animal Performance and Carcass Characteristics of Feedlot Cattle. Ph.D. Thesis, Texas Tech University, Lubbock, TX, USA, May 2020. Available online: https://hdl.handle.net/2346/85832 (accessed on 24 February 2022).
- Stephens, T.P.; Loneragan, G.H.; Karunasena, E.; Brashears, M.M. Reduction of Escherichia coli O157 and Salmonella in feces and on hides of feedlot cattle using various doses of a direct-fed microbial. J. Food Prot. 2007, 70, 2386–2391. [Google Scholar] [CrossRef]
- Younts-Dahl, S.M.; Galyean, M.L.; Loneragan, G.H.; Elam, N.A.; Brashears, M.M. Dietary supplementation with Lactobacillus- and Propionibacterium-based direct-fed microbials and prevalence of Escherichia coli O157 in beef feedlot cattle and on hides at harvest. J. Food Prot. 2004, 67, 889–893. [Google Scholar] [CrossRef] [PubMed]
- Gragg, S.E.; Loneragan, G.H.; Brashears, M.M.; Arthur, T.M.; Bosilevac, J.M.; Kalchayanand, N.; Wang, R.; Schmidt, J.W.; Brooks, J.C.; Shackelford, S.D.; et al. Cross-sectional study examining Salmonella enterica carriage in subiliac lymph nodes of cull and feedlot cattle at harvest. Foodborne Pathog. Dis. 2013, 10, 368–374. [Google Scholar] [CrossRef] [Green Version]
- Arthur, T.M.; Brichta-Harhay, D.M.; Bosilevac, J.M.; Guerini, M.N.; Kalchayanand, N.; Wells, J.E.; Shackelford, S.D.; Wheeler, T.L.; Koohmaraie, M. Prevalence and characterization of Salmonella in bovine lymph nodes potentially destined for use in ground beef. J. Food Prot. 2008, 71, 1685–1688. [Google Scholar] [CrossRef] [PubMed]
- Webb, H.E.; Brichta-Harhay, D.M.; Brashears, M.M.; Nightingale, K.K.; Arthur, T.M.; Bosilevac, J.M.; Kalchayanand, N.; Schmidt, J.W.; Wang, R.; Granier, S.A.; et al. Salmonella in peripheral lymph nodes of healthy cattle at slaughter. Front. Microbiol. 2017, 8, 2214. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention (CDC). Salmonella. Available online: https://www.cdc.gov/salmonella/index.html (accessed on 16 July 2022).
- Laufer, A.S.; Grass, J.; Holt, K.; Whichard, J.M.; Griffin, P.M.; Gould, L.H. Outbreaks of Salmonella infections attributed to beef—United States, 1973–2011. Epidemiol. Infect. 2015, 143, 2003–2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Center for Emerging and Zoonotic Infections. Salmonella and Ground Beef. Available online: https://www.cdc.gov/ncezid/dfwed/prevention-priorities/salmonella-and-ground-beef.html (accessed on 24 February 2022).
- Food Safety Inspection Service—U.S. Department of Agriculture. Public Health Effects of Performance Standards for Ground Beef and Beef Manufacturing Trimmings. 2019. Available online: https://www.fsis.usda.gov/sites/default/files/media_file/2020-08/beef-ps-aug-2019.pdf (accessed on 12 July 2022).
- Vipham, J.L.; Loneragan, G.H.; Guillen, L.M.; Brooks, J.C.; Johnson, B.J.; Pond, A.; Pond, N.; Brashears, M.M. Reduced burden on Salmonella enterica in bovine subiliac lymph nodes associated with administration of direct-fed microbial. Zoonoses Public Health 2015, 62, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Chavez, B.D.; Brashears, M.M. Applications and safety considerations of Lactobacillus salivarius as a probiotic in animal and human health. J. Appl. Microbiol. 2017, 123, 18–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayala, D.I.; Cook, P.W.; Franco, J.G.; Bugarel, M.; Kottapalli, K.R.; Loneragan, G.H.; Brashears, M.M.; Nightingale, K.K. A systematic approach to identify and characterize the effectiveness and safety of novel probiotic strains to control foodborne pathogens. Front. Microbiol. 2019, 10, 1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brashears, M.M.; Nightingale, K.K.; Campos, D. Reduction of Pathogens and Other Bacteria in Food and Feed Products Utilizing a Multiple Inhibition System with Lactic Acid Bacteria. U.S. Patent WO 2019/113217 A1, 13 June 2019. Available online: https://patentimages.storage.googleapis.com/1a/72/ae/c4386992ded080/WO2019113217A1.pdf (accessed on 23 July 2022).
- Bordonaro, R.; McDonough, P.L.; Chang, Y.; Mohammed, H.O. Molecular detection of Salmonella species in bovine fecal samples. J. Vet. Diagn. 2013, 25, 756–758. [Google Scholar] [CrossRef] [Green Version]
- U.S. Food and Drug Administration. BAM R11: Butterfield’s Phosphate-Buffered Dilution Water. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-r11-butterfields-phosphate-buffered-dilution-water (accessed on 9 November 2022).
- Hygiena. BAX® System Real-Time PCR Assay: Salmonella. Available online: https://www.hygiena.com/wp-content/uploads/2020/09/BAX-Q7-Assay-Kit-Insert-Salmonella-RT-English.pdf (accessed on 17 July 2022).
- Liang, K.Y.; Zeger, S.L. Longitudinal data analysis using generalized linear models. Biomet. 1986, 73, 13–22. [Google Scholar] [CrossRef]
- Hanley, J.A.; Negassa, A.; Edwardes, M.D.B.; Forrester, J.E. Statistical analysis of correlated data using generalized estimating equations: An orientation. Am. J. Epidemiol. 2003, 157, 364–375. [Google Scholar] [CrossRef]
- Ballinger, G.A. Using generalized estimating equations for longitudinal data analysis. Organ. Res. Methods 2004, 7, 127–150. [Google Scholar] [CrossRef]
- Clopper, C.J.; Pearson, E.S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 1934, 26, 404–413. [Google Scholar] [CrossRef]
- Tabe, E.S.; Oloya, J.; Doetkott, D.K.; Uer, M.L.; Gib, P.S.; Khaitsa, M.L. Comparative effect of direct-fed microbials on fecal shedding of Escherichia coli O157:H7 and Salmonella in naturally infected feedlot cattle. J. Food Prot. 2008, 71, 539–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Younts-Dahl, S.M.; Osbo, G.D.; Galyean, M.L.; Rivera, J.D.; Loneragan, G.H.; Brashears, M.M. Reduction of Escherichia coli O157 in finishing beef cattle by various doses of Lactobacillus acidophilus in direct-fed microbials. J. Food Prot. 2016, 68, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Wisener, L.V.; Sargeant, J.M.; O’Connor, A.M.; Faires, M.C.; Glass-Kaastra, S.K. The use of direct-fed microbials to reduce shedding of Escherichia coli O157 in beef cattle: A systematic review and meta-analysis. Zoonoses Public Health 2015, 62, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Brashears, M.M.; Galyean, M.L.; Loneragan, G.H.; Mann, J.E.; Killinger-Mann, K. Prevalence of Escherichia coli O157:H7 and performance by beef feedlot cattle given Lactobacillus direct-fed microbials. J. Food Prot. 2016, 66, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Uzal, F.A.; Freedman, J.C.; Shrestha, A.; Theoret, J.R.; Garcia, J.; Awad, M.M.; Adams, V.; Moore, R.J.; Rood, J.I.; McClane, B.A. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol. 2014, 9, 361–377. [Google Scholar] [CrossRef] [Green Version]
- Schoster, T.P.; Loneragan, G.H.; Karunuasena, E.; Brashears, M.M. In vitro inhibition of Clostridium difficile and Clostridium perfringens by commercial probiotic strains. Anaerobe 2013, 20, 36–41. [Google Scholar] [CrossRef]
- Golić, N.; Veljović, K.; Popović, N.; Djokić, J.; Strahinić, I.; Mrvaljević, I.; Terzić-Vidojević, A. In vitro and in vivo antagonistic activity of new probiotic culture against Clostridium difficile and Clostridium perfringens. BMC Microbiol. 2017, 17, 108. [Google Scholar] [CrossRef] [Green Version]
- McReynolds, J.; Waneck, C.; Byrd, J.; Genovese, K.; Duke, S.; Nisbet, D. Efficacy of multistrain direct-fed microbial and phytogenetic products in reducing necrotic enteritis in commercial broilers. Poultry Sci. 2009, 88, 2075–2080. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.R.; Edrington, T.S.; Genovese, K.J.; He, H.L.; Anderson, R.C.; Nisbet, D.J. Evaluation of the efficacy of three direct fed microbial cocktails to reduce fecal shedding of Escherichia coli O157:H7 in naturally colonized cattle and fecal shedding and peripheral lymph node carriage of Salmonella in experimentally infected cattle. J. Food Prot. 2019, 83, 28–36. [Google Scholar] [CrossRef] [PubMed]


| Date | Treatment | Number of Samples | ||
|---|---|---|---|---|
| Pen 1 | Pen 2 | Pen 3 | ||
| 26-May-2021 | Control | 10 | 10 | 2 |
| 28-Jun-2021 | Control | 10 | 10 | 2 |
| 27-Jul-2021 | Control | 10 | 10 | 2 |
| 23-Aug-2021 | Control | 10 | 10 | 2 |
| 21-Sep-2021 | Control | 10 | 10 | 2 |
| 26-May-2021 | BD | 5 | 5 | 5 |
| 28-Jun-2021 | BD | 5 | 5 | 5 |
| 27-Jul-2021 | BD | 5 | 5 | 5 |
| 23-Aug-2021 | BD | 5 | 5 | 5 |
| 21-Sep-2021 | BD | 5 | 5 | 5 |
| 26-May-2021 | PC | 10 | 10 | 3 |
| 28-Jun-2021 | PC | 10 | 10 | 3 |
| 27-Jul-2021 | PC | 10 | 10 | 3 |
| 23-Aug-2021 | PC | 10 | 10 | 3 |
| 21-Sep-2021 | PC | 10 | 10 | 3 |
| Term | Estimate (lnOR) | Standard Error | p-Value |
|---|---|---|---|
| Intercept | −1.92 | 0.60 | 0.001 |
| Probicon | −0.88 | 0.43 | 0.039 |
| Bovamine Defend | −0.85 | 0.52 | 0.099 |
| June 2021 | 1.33 | 0.64 | 0.039 |
| July 2021 | −1.69 | 1.01 | 0.093 |
| August 2021 | −0.98 | 1.02 | 0.338 |
| September 2021 | 1.67 | 0.66 | 0.012 |
| Term | Estimate (lnOR) | Standard Error | p-Value |
|---|---|---|---|
| Intercept | −1.64 | 0.31 | <0.001 |
| Probicon | −3.05 | 0.68 | <0.001 |
| Bovamine Defend | −2.61 | 0.73 | <0.001 |
| June 2021 | 1.76 | 0.43 | <0.001 |
| July 2021 | 0.00 | 0.90 | 1.000 |
| August 2021 | 0.00 | 0.90 | 1.000 |
| September 2021 | 0.00 | 0.70 | 1.000 |
| Term | Estimate | Standard Error | p-Value |
|---|---|---|---|
| Intercept | 1.97 | 0.29 | <0.001 |
| Probicon | −1.08 | 0.28 | <0.001 |
| Bovamine Defend | 0.06 | 0.26 | 0.822 |
| July 2021 | 3.23 | 0.35 | <0.001 |
| August 2021 | 0.66 | 0.37 | 0.071 |
| September 2021 | 1.77 | 0.32 | <0.001 |
| Term | Estimate | Standard Error | p-Value |
|---|---|---|---|
| Intercept | 6.16 | 0.26 | <0.001 |
| Probicon | 0.12 | 0.27 | 0.65 |
| Bovamine Defend | 0.45 | 0.28 | 0.11 |
| May 2021 | −0.42 | 0.28 | 0.13 |
| July 2021 | −1.30 | 0.49 | 0.007 |
| September 2021 | −1.07 | 0.27 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flach, M.G.; Dogan, O.B.; Kreikemeier, W.M.; Nightingale, K.K.; Brashears, M.M. Reduction of Pathogens in Feces and Lymph Nodes Collected from Beef Cattle Fed Lactobacillus salivarius (L28), Lactobacillus acidophilus (NP51) and Propionibacterium freudenreichii (NP28), Commercially Available Direct-Fed Microbials. Foods 2022, 11, 3834. https://doi.org/10.3390/foods11233834
Flach MG, Dogan OB, Kreikemeier WM, Nightingale KK, Brashears MM. Reduction of Pathogens in Feces and Lymph Nodes Collected from Beef Cattle Fed Lactobacillus salivarius (L28), Lactobacillus acidophilus (NP51) and Propionibacterium freudenreichii (NP28), Commercially Available Direct-Fed Microbials. Foods. 2022; 11(23):3834. https://doi.org/10.3390/foods11233834
Chicago/Turabian StyleFlach, Makenzie G., Onay B. Dogan, Wanda M. Kreikemeier, Kendra K. Nightingale, and Mindy M. Brashears. 2022. "Reduction of Pathogens in Feces and Lymph Nodes Collected from Beef Cattle Fed Lactobacillus salivarius (L28), Lactobacillus acidophilus (NP51) and Propionibacterium freudenreichii (NP28), Commercially Available Direct-Fed Microbials" Foods 11, no. 23: 3834. https://doi.org/10.3390/foods11233834
APA StyleFlach, M. G., Dogan, O. B., Kreikemeier, W. M., Nightingale, K. K., & Brashears, M. M. (2022). Reduction of Pathogens in Feces and Lymph Nodes Collected from Beef Cattle Fed Lactobacillus salivarius (L28), Lactobacillus acidophilus (NP51) and Propionibacterium freudenreichii (NP28), Commercially Available Direct-Fed Microbials. Foods, 11(23), 3834. https://doi.org/10.3390/foods11233834






